FIGURE 1.
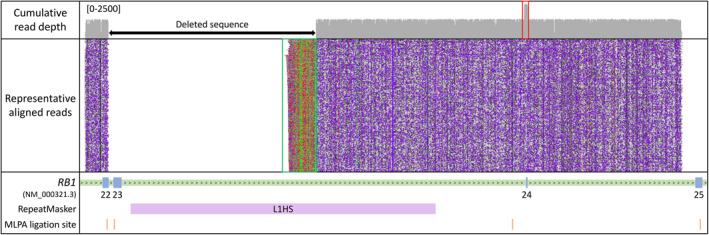
Representative long‐read sequencing alignments following long‐range polymerase chain reaction enrichment of the RB1 target locus. Variant‐containing reads were size selected, in silico, to aid identification of the deletion breakpoint, which intersects an L1HS long interspersed nuclear element at its 3′ end. “Soft‐clipping” of the reads indicates the presence of an 85 bp insertion (see green box which corresponds to the non‐aligning portion of the read), for which 81 bp correspond to the downstream exon 24 locus (see red box which corresponds to increased read‐depth). Multiplex ligation‐dependent probe amplification (MLPA) ligation sites are marked. It is notable that the exon 24 probe is sited 265‐bp upstream of the exon boundary (and outside the duplicated segment). The y‐axis scale for the cumulative read‐depth plot is labelled. Arrows denote the direction of transcription
