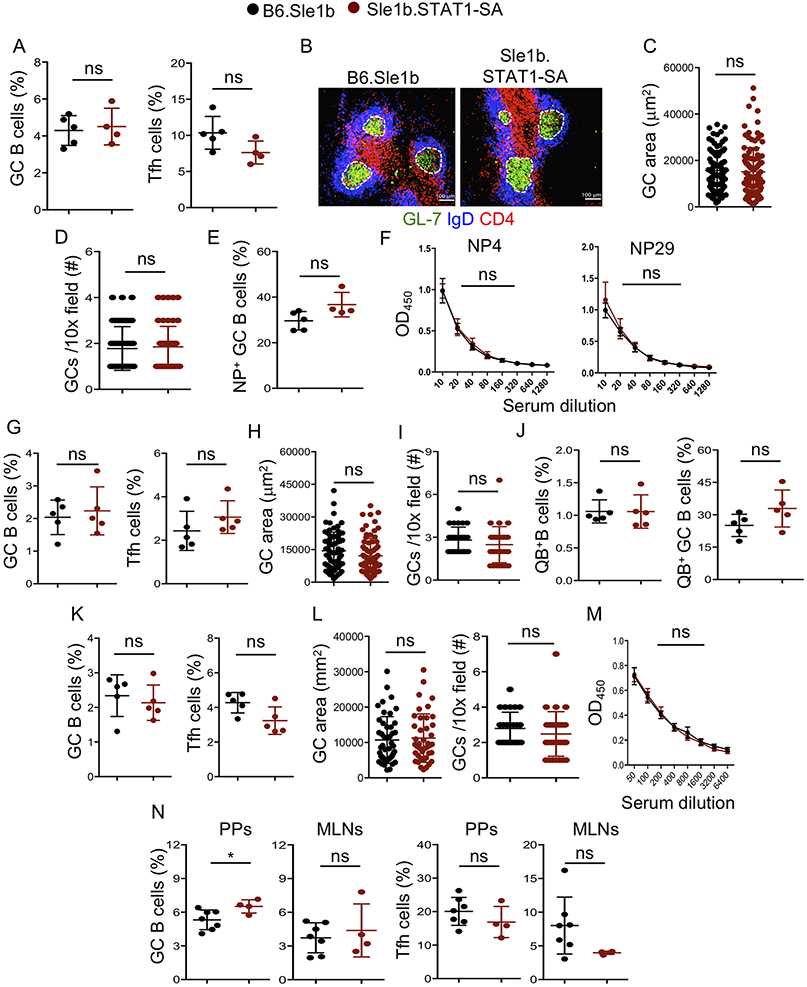
Figure 3. STAT1-pS727 deficiency does not alter immune response to T dependent antigens, VLPs and viral infection.
(A-F) These data were generated from spleens of NP-KLH immunized B6.Sle1b and Sle1b.STAT1-SA female mice. (A) Quantification of B220+GL7hiCD95hi GC B cells and CD4+CD44hiPD-1hiCXCR5hi Tfh cells that were gated on total B220+ B and CD4+ T cells, respectively. Representative histological images show GCs (B), GC area (C) and GC frequency (D) in spleen sections. (E) Percentage of NP18-specific B220+GL7hiCD95hi GC B cells that were gated on total B220+ B cells. (F) NP4- and NP29-specific IgG titers were measured by ELISA. (G-J) These data were derived from VLP immunized B6.Sle1b and Sle1b.STAT1-SA female mice. (G) Flow cytometry data depicts frequency of splenic B220+GL7hiCD95hi GC B cells and CD4+CD44hiPD-1hiCXCR5hi Tfh cells that were gated on total B220+ B and CD4+ T cells, respectively. Quantification of GC area (H) and GC frequency (I) in spleen sections. (J) Percentage of splenic VLP-specific Qβ+ B and GC B cells. (K-M) These data were derived from mouse polyoma virus (mPyV) infected B6.Sle1b and Sle1b.STAT1-SA female mice. (K) Frequency of splenic B220+GL7hiCD95hi GC B cells and CD4+CD44hiPD-1hiCXCR5hi Tfh cells. (L) Quantification of GC area and GC frequency in spleen sections. (M) VP1 specific IgG titers measured by ELISA. (N) Quantitation of GC B cells and Tfh cells from total B220+ B and CD4+ T cells, respectively, in Peyer’s patches (PPs) and mesenteric lymph nodes (MLNs) in unimmunized mice. These data represent 2-3 experiments and each symbol indicates an individual mouse or GC. 3-5 mice were analyzed in each experiment. Statistical analysis was performed by two-way ANOVA, with a follow-up Sidak multiple-comparison test (F, M) or unpaired, nonparametric Mann-Whitney Student’s t-test. ns, non-significant; *, P < 0.05.