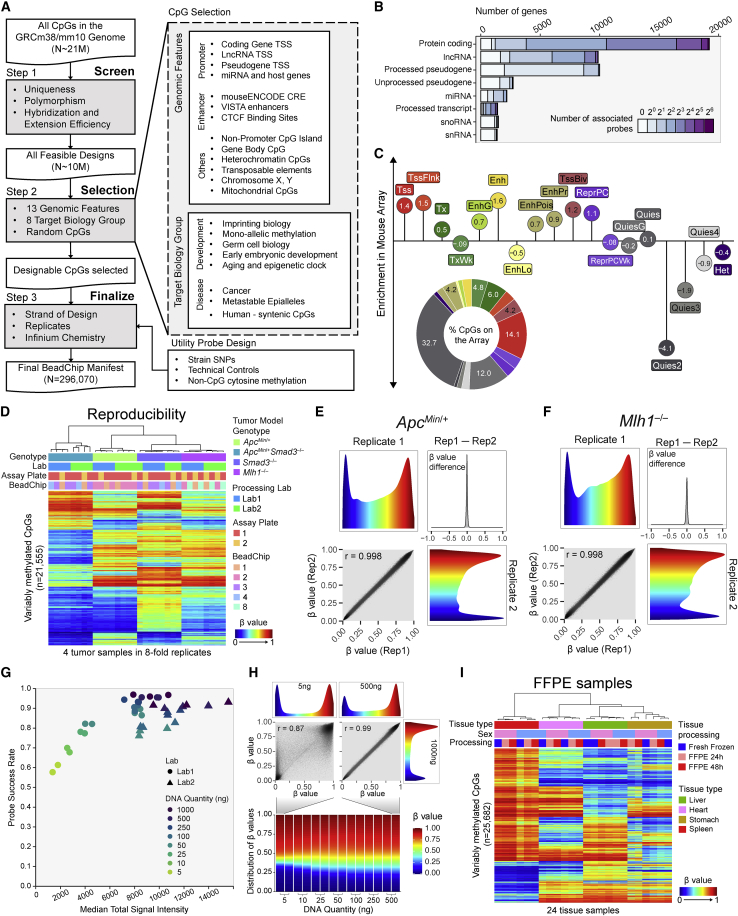
Figure 1.
Mouse DNA methylation array design and technical validation
(A) Workflow of designing the mouse Infinium methylation BeadChip.
(B) The number of probes associated with each gene model biotype.
(C) Enrichment of the probes in different chromatin states. The consensus chromatin states from 66 ENCODE ChromHMM files for twelve tissues were used. Distribution of interrogated CpGs in different chromatin states is shown at bottom left.
(D) Heatmap representing unsupervised clustering of four intestinal tumor samples, each analyzed in 8-fold replicate, including at two separate laboratory facilities. 21,643 most variable probes are shown.
(E and F) Smooth scatterplot of two replicates from ApcMin (E) or Mlh1 (F) tumor samples. The colors represent DNA methylation β values, and the β value difference between the two replicates is shown on the top right quadrant.
(G) Probe success rate of samples of DNA input ranging from 5 to 1,000 ng. Data from two labs are represented by different shapes.
(H) Smooth scatterplots contrasting the comparison of a low-input sample (5 ng) and a high-input sample (500 ng), both with 1,000-ng-input sample. Samples are losing intermediate methylation reading as DNA input decreases.
(I) Reproducibility of DNA methylation measurement from four tissues comparing fresh frozen samples and samples fixed with formalin for 24 and 48 h.
See also Figures S1 and S2; Table S1.