Figure 4.
Identification of tissue-specific DNA methylation patterns and signatures
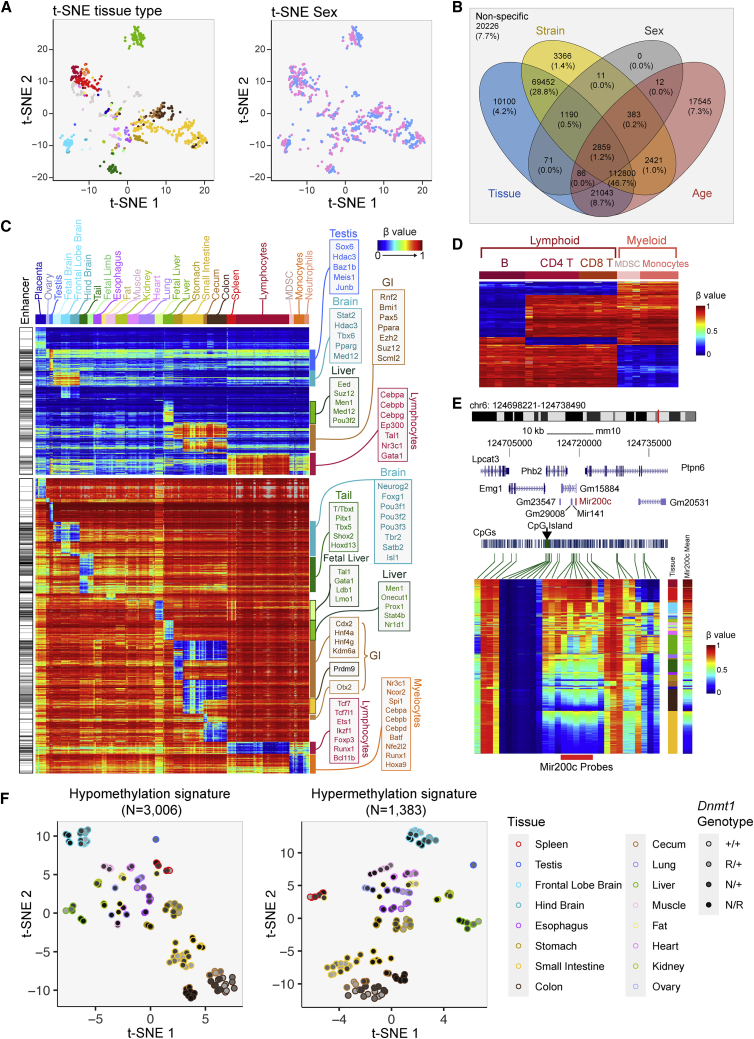
(A) t-SNE cluster map showing samples clustered by tissue type (left) and sex (right). The color legend is the same as in Figure 3A.
(B) Four-way Venn diagram showing CpGs differentially methylated with respect to strain, tissue, sex, and age in 467 mouse samples.
(C) CpGs (rows) specifically methylated (top) and unmethylated (bottom) in each tissue (columns showing samples organized by tissue type).
(D) CpGs (rows) specifically methylated (top) and unmethylated (bottom) in each sorted leukocyte cell type (columns showing samples organized by cell type).
(E) Track view of the DNA methylation profile of the Mir200c locus as a marker for epithelial versus mesenchymal cells. Tissue type and mean methylation level of five CpGs associated with the Mir200c locus (shown by the red bar at the bottom) are shown on the right-hand side of the heatmap.
(F) t-SNE cluster map of primary tissue samples from wild-type and Dnmt1NR mice using tissue-specific hypomethylation (left) and hypermethylation signature (right). The color legend is the same as in Figure 3A.
See also Figure S5.