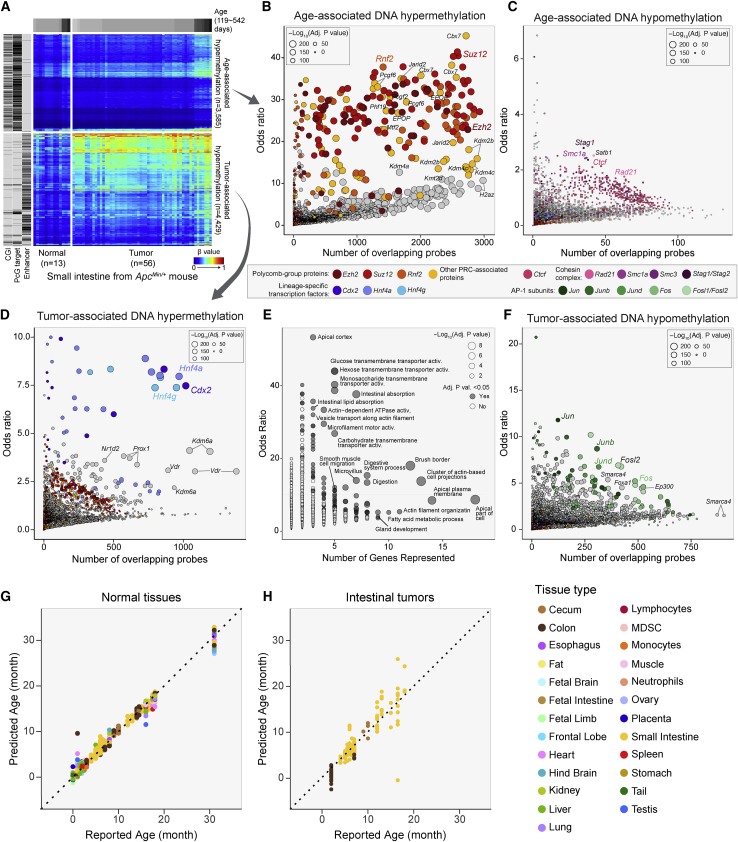
Figure 7.
Age- and tumor-associated DNA methylation and the development of an epigenetic clock
(A) Heatmap displaying DNA methylation gain associated with aging (top) and with tumorigenesis (bottom). Rows correspond to CpGs and columns correspond to samples. Left-hand side bar represents whether probes (rows) fall to CpG islands (CGI), Polycomb repressive complex group (PcG) target, and enhancer elements in small intestine samples.
(B) Transcription factors whose binding sites are enriched in CpGs that gain methylation with age. y axis represents odds ratio of enrichment, and x axis represents number of significant probes overlapping binding sites. Size of the dots denotes statistical significance of the enrichment (Fisher’s exact test).
(C) Transcription factors whose binding sites are enriched in CpGs that lose methylation with age.
(D) Transcription factors whose binding sites are enriched in CpGs that gain methylation associated with tumorigenesis. Each dot corresponds to a transcription factor.
(E) Ontology terms enriched in target genes regulated by enhancers that gain DNA methylation.
(F) Transcription factors whose binding sites are enriched in CpGs that lose methylation associated with tumorigenesis.
(G) Scatterplot comparing age predicted by a mouse epigenetic clock using 347 CpG probes (y axis) and reported age (x axis) in disease-free tissues.
(H) Scatterplot comparing age predicted by a mouse epigenetic clock using 347 CpG probes (y axis) and reported age (x axis) in mouse intestinal tumors.