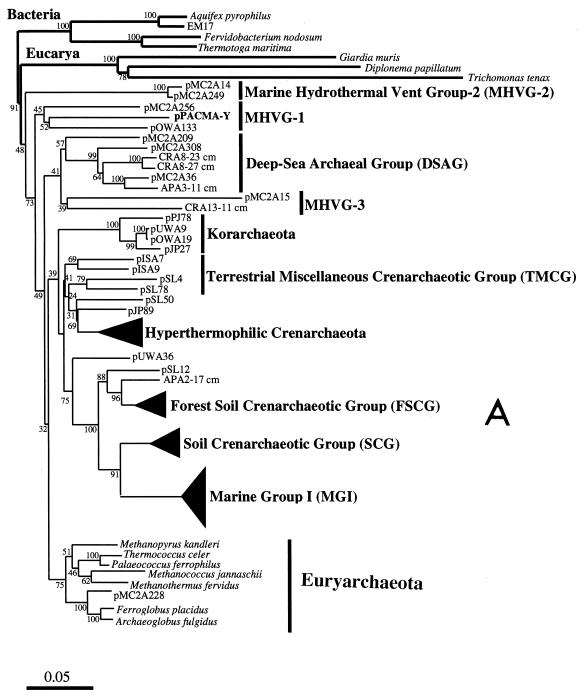
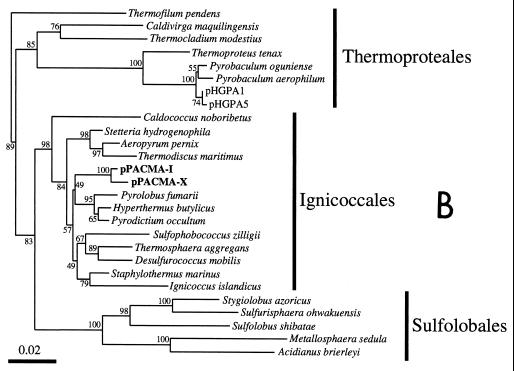
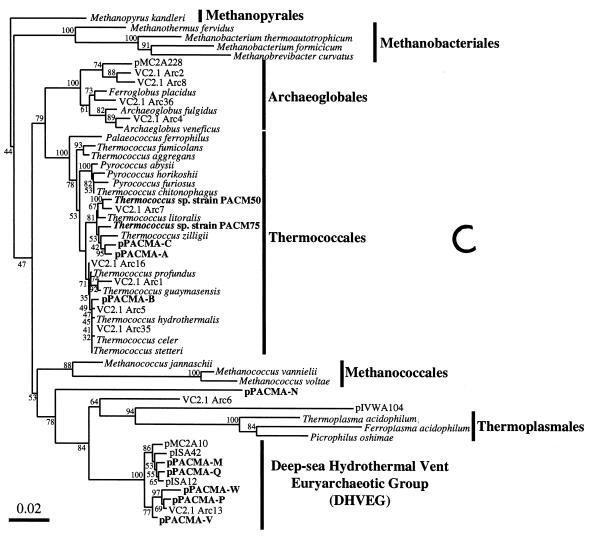
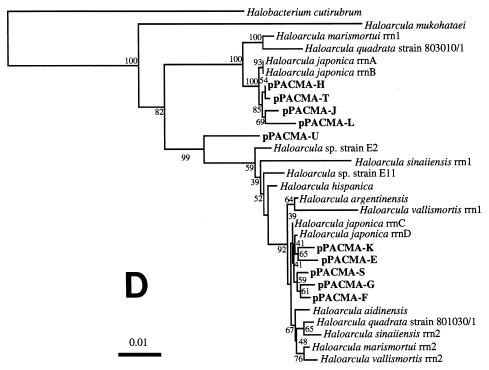
FIG. 3.
Phylogenetic trees based on SSU rDNA sequences including various rDNA clones obtained from the black smoker chimney at the PACMANUS site in the Manus Basin. Each of the trees was inferred by neighbor-joining analysis of approximately 600 homologous positions of the rDNA sequence. (A) A tree indicating the phylogenetic relationship among the domains of Bacteria and Eucarya, the deep branches of uncultivated archaea, and the crenarchaeotic and euryarchaeotic kingdoms. (B) A tree indicating the phylogenetic organization among the hyperthermophilic crenarchaeota, Thermoproteales, Igniococcales, and Sulfolobales. (C) A tree indicating the phylogenetic relationship within the hyperthermophilic and thermophilic euryarchaeota and the possible thermophilic euryarchaeotic phylotypes. (D) A tree indicating the phylogenetic organization within the genus Haloarcula. The numbers on the branches represent the bootstrap confidence values. The scale bars indicate the expected changes per sequence position. Abbreviations indicate rDNA clones corresponding to uncultivated organisms derived from the following environments: pMC1A, pMC2A, pISA, and pIVWA from deep-sea hydrothermal vent environments (51); pOWA and pUWA from shallow marine hydrothermal vent water and terrestrial acidic hot spring water, respectively (55); pJP and pSL from sediments in Yellowstone National Park hot springs (3, 4); CRA and APA from deep-sea sediments (59); pHGPA from deep subsurface geothermal water (50); VC2.1 Arc from an in situ growth chamber (Vent Cap) deployed at a deep-sea hydrothermal vent (44); and pPACMA from a black smoker chimney at the PACMANUS site in the Manus Basin.