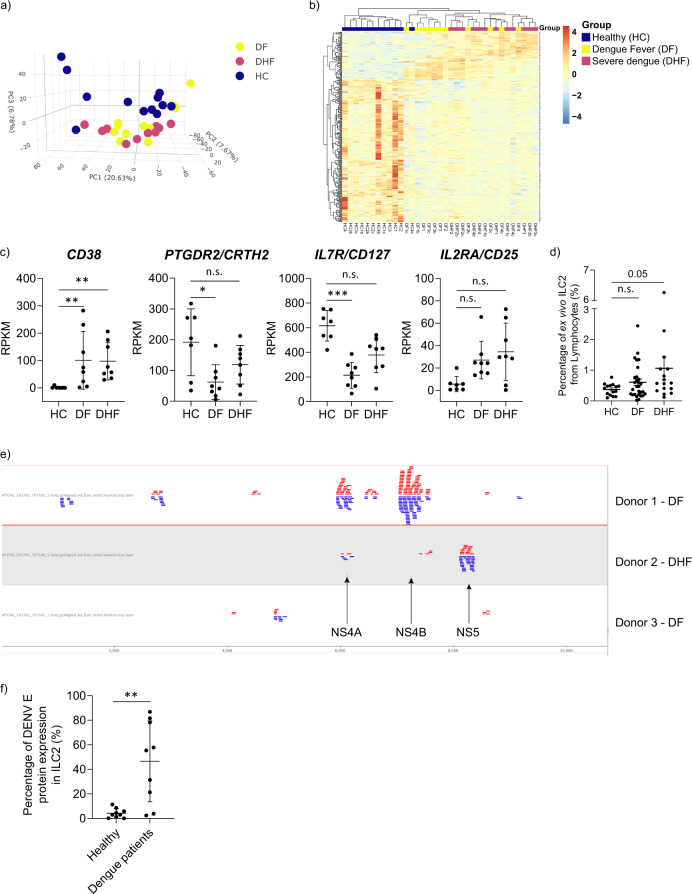
Fig. 1. In vivo ILC2 are activated and infected in dengue infection.
a Principal component analysis and b heat map of DEGs (Likelihood ratio test, FDR < 0.05) from ex vivo ILC2 from three separate groups of individuals: healthy individuals (HC, navy blue) (n = 3), Dengue Fever (DF, yellow) (n = 4) and Dengue Haemorrhagic Fever (DHF, violet) (n = 4) in duplicates. DF and DHF groups are categorised using WHO SEARO classification. c Activation related gene expression of CD38, PTGDR2 (CRTH2), CD127(IL-7Rα), IL-2RA (CD25) of human blood-derived ILC2 of HC (n = 3), DF (n = 4) and DHF (n = 4), determined by RNA Sequencing and represented in RPKM values. Statistical analysis performed using adjusted p value from the DEG result. P * < 0.05, ** <0.01, *** <0.001, n.s. not significant. d Frequency of ILC2 in healthy individuals (HC) (n = 15), Dengue Fever (DF)(n = 29) and Dengue Haemorrhagic Fever (DHF) (n = 16). ILC2 were stained as lineage-, CD45+, CD3-, CRTH2+, CD127+ (Statistical significance was tested using one-way ANOVA with Tukey’s multiple comparison test. P 0.05, n.s. not significant.). e Mapping figure from genome browser depicting matching genes of dengue genome to the raw RNA sequencing data from ILC2 in patients having Dengue fever (n = 2) and Dengue Haemorrhagic Fever (n = 1). Corresponding proteins for the base pair ranges were depicted. 6376~6756 bp: Non-Structural protein NS4A, 6757–6825 bp: protein 2K, 6826–7569 bp: Non-Structural protein NS4B, 7570~10269 bp: RNA-dependent RNA polymerase NS5. f For staining of dengue infection of ILC2, PBMC from HC (n = 9) and patients with acute dengue infection (n = 9) from Sri Lankan ethnicity were used. Cryopreserved PBMC samples were thawed and stained for ILC2 (Live/CD45+/lineage/CD3/CD56-/CRTH2+/IL7Rα+) and for intracellular dengue virus envelope (E) protein. Dengue infection was represented as percentage from total ILC2 (Statistical significance was tested using T-test. P * < 0.05, ** <0.01, *** <0.001). All error bars represent mean ± SD.