Abstract
Roots are the interface between the plant and the soil and play a central role in multiple ecosystem processes. With intensification of agricultural practices, rhizosphere processes are being disrupted and are causing degradation of the physical, chemical and biotic properties of soil. However, cover crops, a group of plants that provide ecosystem services, can be utilised during fallow periods or used as an intercrop to restore soil health. The effectiveness of ecosystem services provided by cover crops varies widely as very little breeding has occurred in these species. Improvement of ecosystem service performance is rarely considered as a breeding trait due to the complexities and challenges of belowground evaluation. Advancements in root phenotyping and genetic tools are critical in accelerating ecosystem service improvement in cover crops. In this study, we provide an overview of the range of belowground ecosystem services provided by cover crop roots: (1) soil structural remediation, (2) capture of soil resources and (3) maintenance of the rhizosphere and building of organic matter content. Based on the ecosystem services described, we outline current and promising phenotyping technologies and breeding strategies in cover crops that can enhance agricultural sustainability through improvement of root traits.
Keywords: exudation, genetic selection, nitrogen fixation, polyculture, resource capture, root phenotyping, soil compaction, soil organic matter
Summary
Ecosystem service performance is rarely considered as a breeding trait due to the complexities and challenges of belowground evaluation. Advancements in phenotyping technologies and breeding strategies in cover crops are critical to agricultural sustainability through improvement of root traits.
1. INTRODUCTION
The world population is currently sustained by food produced on one‐third of the world land mass. Population levels are increasing every year with food demand expected to outstrip production by 2050. Total arable land use worldwide has decreased in the last decade; however, there is a continued pressure to expand agricultural land use to meet rising food demand (FAO, 2021). To meet this demand, agricultural intensification practices such as chemical fertiliser and herbicide inputs have enabled greater food productivity per unit area (Bommarco et al., 2013). At present these inputs are cost effective for maximising yield, but they are not sustainable as they are dependent on fossil fuels. Intensification of agriculture also has hidden costs on the soil, such as reducing soil organic carbon, decreasing insect and microbial diversity, and increasing soil compaction by heavy machinery; each of which can eventually reduce yield (Gurr et al., 2016; Kaspar & Singer, 2011). A shift to sustainable intensification practices that incorporate ecological service into management and breeding are needed to protect and restore soil and to reduce our dependence on chemical fertilisers and herbicides, whilst maintaining or improving crop yields (Tittonell, 2014).
Fallow periods in cropping systems, where the soil is left bare (typically during the cool season months), are a common practice in modern intensive agriculture such as Western Europe and the US‐Midwest. Planting and tilling practices leave the soil exposed from harvest through spring. During the off‐season a wide group of plant species, known as cover crops, can be utilised to cover the bare ground during this time (Hallama et al., 2019). Having plants growing in a field all year round and reducing fallow spaces in cropping systems help to maintain ecological processes which otherwise would be disrupted (Magdoff & van Es, 2009). Cover crops are primarily used to provide ecosystem functions in the field and do not necessarily have a direct economic output or harvestable product. Although there are emerging exceptions such as alfalfa that provides forage and pennycress that produces high‐quality oilseeds in addition to ecosystem services (McGinn et al., 2019; Santos et al., 2011). Cover crops have also been used to help close the yield gap between conventional and organic farming (Hansen et al., 2021). Cover crop species are mostly found across the grasses, brassicas and legume families with the number of proposed species increasing rapidly. Each of these cover crop families provide a variety of ecosystem functions (Table 1). The fibrous root systems of grasses are particularly effective for nutrient capture while the tap root systems of brassicas are effective at deep rooting and breaking up compacted soil layers through root swelling. Legumes are able to form root nodules that provide biological nitrogen fixation through a symbiotic relationship with rhizobia.
Table 1.
Primary ecosystem functions provided by roots of cover crop species
| Family | Name | Soil erosion protection | Increased SOM content | Nutrient and water 'catch' | Topsoil structure remediation | Subsoil structure remediation | Soil pathogen resistance | Biological N fixation | Notes |
|---|---|---|---|---|---|---|---|---|---|
| Cool‐season annual grains | |||||||||
| Poaceae | Barley (Hordeum vulgare) | x | x | x | x | x | x | Planted in spring or fall, commonly mixed with peas, oats, and crimson or red clover | |
| Oat (Avena sativa) | x | x | x | x | x | Planted in spring or fall, commonly mixed with alfalfa, hairy vetch and field peas | |||
| Wheat, triticale, spelt (Triticum spp.) | x | x | x | x | x | Planted in spring or fall, commonly mixed with peas, vetch, and clover | |||
| Winter rye (Secale cereale) | x | x | x | x | x | Planted in fall | |||
| Warm‐season annual grains | |||||||||
| Polygonaceae | Buckwheat (Fagopyrum esculentum) | x | x | x | x | Planted in spring through summer | |||
| Sudangrass, sorghum‐sudangrass (Sorghum bicolor × S. bicolor var Sudanese) | x | x | x | x | x | x | Planted in spring through summer, mix with a buckwheat crop or with forage soybeans and cowpeas | ||
| Japanese millet (Echinochloa esculenta), pearl millet (Pennisetum glaucum), foxtail millet (Setaria italica) | x | x | x | x | Planted in spring through summer, commonly mixed with forage soybeans and cowpeas | ||||
| Teff (Eragrostis tef) | x | x | x | x | Planted in spring through summer | ||||
| Brassicas | |||||||||
| Brassicaceae | Mustard, canola (Brassica spp.) | x | x | x | x | x | Planted in spring or fall | ||
| Arugula (Eruca sativa) | x | x | x | x | x | Planted in fall | |||
| Pennycress (Thlaspi arvense) | x | x | x | x | Planted in spring or fall | ||||
| Forage radish (Raphanus sativus), forage turnip (Brassica rapa) | x | x | x | x | x | Planted in fall | |||
| Grasses | |||||||||
| Annual ryegrass, perennial ryegrass (Lolium spp.) | x | x | x | x | x | Planted in spring or fall | |||
| Orchardgrass (Dactylis glomerata), timothy (Phleum pratense) | x | x | x | x | Planted in spring or fall | ||||
| Legumes | |||||||||
| Fabaceae | Field pea (Pisum sativum) | x | x | x | x | x | x | Planted in spring or fall | |
| Soybean (Glycine max), cowpea (Vigna unguiculata) | x | x | x | x | x | x | Planted in spring through summer | ||
| Alfalfa (Medicago sativa) | x | x | x | x | x | Planted in spring or fall, is best grown with a small grain crop or perennial grass | |||
| Hairy vetch (Vicia villosa) | x | x | x | x | x | x | x | Planted in fall, commonly planted with winter cereals. | |
| Chickling vetch (Lathyrus sativus) | x | x | x | x | x | Planted in spring through summer | |||
| Perennial clovers. Red clover, white clover, alsike clover (Trifolium spp.), yellow sweetclover (Melilotus officinalis) | x | x | x | x | x | x | Planted in spring or fall | ||
| Annual clovers. Crimson clover, berseem clover, subterranean clover (Trifolium spp.) | x | x | x | x | x | x | Planted in spring or fall | ||
Note: Red colour indicates high performance for a particular ecosystem service. Adapted from NRCS Cover Crop plant specification guide: https://www.nrcs.usda.gov/Internet/FSE_DOCUMENTS/stelprdb1081555.pdf
Introducing cover cropping into conventional crop rotation systems provides economic and ecological benefits over multiple years. Ecosystem benefits of using cover crops include increasing biodiversity, increasing numbers of pollinators, increasing soil organic carbon content, improving soil fertility and reducing soil erosion (Abdalla et al., 2019; Eberle et al., 2015; Langdale et al., 1991) (Figure 1). Specific cover crops can also provide agrosystem services such as weed suppression and biofumigation for reducing soil pests and pathogens (Henderson et al., 2009; Osipitan et al., 2019). Adoption of cover crops in agriculture is a promising approach toward the vision of sustainable intensification of agriculture (Hunter et al., 2017). The ecosystem services provided by cover crops across multiple years could help to maintain or increase subsequent cash crop yields while using fewer chemical fertilisers and herbicides (Wittwer & van der Heijden, 2020). However, the effectiveness of cover cropping requires the selection of species appropriate to the desired ecosystem function and a compatible phenology with the crop season and management strategy (Osipitan et al., 2019). Yield penalties are common within the first year, and improvement in soil quality and subsequent crop yields are often not observed immediately. Therefore, cover cropping is seen as an investment strategy to maintain and improve soil with observable improvements in subsequent years (Magdoff & van Es, 2009). Unlike, most major cash crops, cover crops have not yet undergone intensive selection and breeding. There is, therefore, a tremendous opportunity for genetic improvements within species used as cover crops in providing more effective ecosystem services to agricultural systems.
Figure 1.
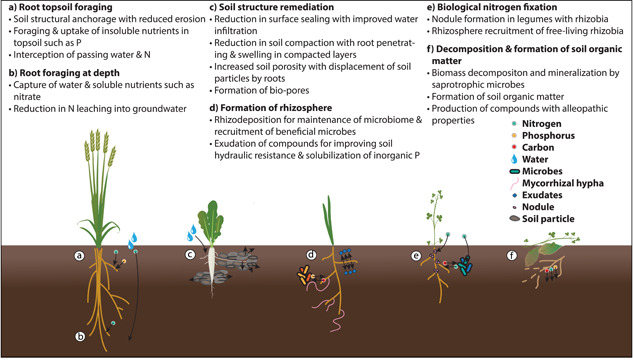
Cover crop root traits and mechanisms of action in providing ecosystem service [Color figure can be viewed at wileyonlinelibrary.com]
Targets in cover crop breeding are likely not the typical yield components valued in cash crop breeding; rather, cover crop breeding will focus on ecosystem services, of which roots are a key component. Cover crop roots play a critical role belowground in resource capture and reducing soil erosion in addition to well‐known above‐ground services (Hallama et al., 2019). The root is the organ for water and nutrient uptake, and roots must establish and arrange themselves in the soil to intercept passing soil resources or grow into new areas to forage for resources (Morris et al., 2017). Roots influence the soil zone around them, termed the rhizosphere; Root growth and exudation affects the physical, chemical and microbial properties of soil (Helliwell et al., 2017; York et al., 2016). In recent years, there has been increased research interest to improve root systems of plants; however, evaluation of root traits remains a technical challenge. Well‐recognised areas for improvement with root systems include abiotic stress tolerance, resource use efficiency, and yield (Reynolds et al., 2021). However, breeding targets need to be more diverse and include evaluation and improvement of root traits related to ecosystem service performance.
In this study, we outline the ecosystem services provided by roots of different families of cover crops. We also provide insights into promising phenotyping and genetic approaches that will aid genetic gains of cover crop root traits. Experimental approaches and insights gained from improving ecosystem service performance will also accelerate our development of similar root traits in cash crops. Understanding root function and influence on the soil environment in cover crop and cash crop systems is important for sustainable agricultural intensification.
2. BELOWGROUND ECOSYSTEM SERVICES PROVIDED BY COVER CROPS
2.1. Soil structure remediation
Soil is a vital living ecosystem that sustains plants, animals, and humans, and soil health reflects the continued capacity of soil in maintaining environmental quality and biodiversity (Bünemann et al., 2018; Lehmann et al., 2020). In agricultural systems, soil health is important in supporting crop productivity and healthy ecosystems. The composition and structure of soil can affect measures of plant resource availability such as water infiltration, water holding capacity, nutrient absorption, nutrient release, gas exchange and root development (Aravena et al., 2014; Carminati et al., 2010; Comerford 2005; Pires et al., 2019; Tracy et al., 2015). Technological advances of the 'Green Revolution' were instrumental in agricultural intensification of cropping systems; however, these technologies are causing the physical, chemical and ecological degradation of soil. Therefore, improvement in soil management is needed to help protect soils and make agriculture more sustainable.
Planting cover crops in bare fallow can greatly reduce soil erosion. Traditional agricultural practices involve fallow periods and tillage; both of which are primary drivers of soil erosion. Without plant cover, soils are susceptible to erosion as topsoil dries between cropping cycles. Rainfall events on bare, hard soil can cause poor water infiltration and high levels of surface runoff which displaces topsoil (Gyssels et al., 2005). Topsoil, water and nutrients are all lost during this process. Roots stabilise soil through structural anchorage and reduce displacement by flowing water. Cover crops usage increased aggregate stability by 1% every year, an important measure of soil erosion resistance (Wood & Bowman, 2021). Grasses are particularly effective for preventing soil erosion as they often produce more root length density in the topsoil compared to taproot species. Shallow and dense rooting plays a role in determining erosion resistance, with increasing root mass leading to decreased water erosion rates (Gyssels et al., 2005). Soil infiltration is also very important with greater infiltration reducing surface runoff and topsoil erosion. At the soil surface, roots displace soil and make channels for water which reduces surface sealing (Y. Liu et al., 2019). Lateral roots also displace soil as they grow, and these roots increase the free space inside and connectivity between soil pores increasing the capacity for water storage and gas exchange (Helliwell et al., 2017). A fallow field being devoid of roots will be more compacted posing a narrower water content range and reduced water flow and will have an increased risk of surface runoff in high rainfall events (Magdoff & van Es, 2009). Breeding and selection of cover crop varieties that have a greater root biomass and exploration in the topsoil is highly desirable for improved water infiltration, soil porosity, and soil stability.
Cover crops can be used during fallow periods to reduce soil compaction and improve cash crop yields in subsequent periods. The use of heavy machinery has been instrumental for intensifying agricultural practices, but mechanisation increases soil compaction. The high mechanical impedance of compacted soils limits root exploration. A size‐limited root system reduces foraging capacity for water and nutrients and in turn, affects plant growth. Root growth in compacted soils is both by poor aeration under high moisture content and impeded by higher soil strength during drier conditions (Magdoff & van Es, 2009). Cover crops can reduce soil compaction by different mechanisms dependent on family type. Both tap‐ and fibrous‐root systems effectively penetrate highly compacted soil layers (Burr‐Hersey et al., 2017). Tap‐rooted plants like forage radish, alfalfa and chicory can penetrate and break up compacted subsoil by root swelling leaving soil biopores (Chen & Weil, 2011; Han et al., 2015). Biopores generated by root growth from previous plantings can induce and facilitate root growth for subsequent crops (Han et al., 2015). Roots of the cash crop following a cover crop will have less impediment to grow deeper by following existing biopores and capture soil resources located in deeper soil layers (Huang et al., 2020; Zhou et al., 2021).
2.2. Soil resource capture
Chemical fertilisers are unsustainable resources that involve manufacturing processes reliant on fossil fuels, and mining of finite reserves such as phosphorus, which are damaging to the environment. However, fertiliser application is a current necessity for agricultural intensification practices with nitrogen (N) and phosphorus (P) often being the limiting nutrients for attaining the maximum yield potential. Maintaining yields with fertiliser application is the highest capital cost for farmers with rising fertiliser prices affecting the price of food for consumers. Lack of accessibility to fertilisers disproportionately affects poor countries since land quality differences between rich and poor countries do not justify the yield gaps (Adamopoulos & Restuccia, 2021). Fertiliser use is also inefficient in intensive agricultural practices with as little as 18%–49% N fertiliser recovery rate across maize, rice and wheat fields (Cassman et al., 2002). Nutrient losses then enter and pollute waterways and groundwater which causes eutrophication and hypoxic zones (Dodds 2006; Malhi et al., 2001).
One strategy to improve soil nutrition and fertiliser use efficiency is with the use of ‘catch’ cover crops (e.g., radish, wheat and sudangrass) (Table 1). During a fallow period, catch cover crops immobilise residual nutrients and are incorporated into plant biomass that are released to subsequent crops after crop termination. Catch cover crops influence nutrient cycling in the field by capturing remnant fertiliser applied to the previous cash crop that otherwise would be lost from the root zone or by scavenging and mobilising P (Heuermann et al., 2019). Nutrient use efficiency is an established target of improvement for cash crops, which consists of uptake and utilisation of a nutrient into the harvestable product. For cover crops, the utilisation of a nutrient by a particular tissue of the plant is often of less importance. Nutrient capture and conversion into plant biomass can reduce nutrient losses in fields. The spatial distribution of roots in time and space, referred to as 'root system architecture' (RSA), determines the exploratory volume of soil where nutrient and water uptake can occur. Key rhizosphere processes, essential for capturing often dynamic and heterogenous soil resources, happen at the root surface (Hallama et al., 2019). Thus, the spatial distribution of root over time is important to consider (Morris et al., 2017). Root development of cover crops can also indirectly influence nutrient cycling with soil structural changes benefiting root exploration of following cash crops through formation of biopores. Biopores that facilitate deeper rooting can uplift limiting elements for plants via nutrient remobilisation and litter fall (Kautz et al., 2013).
For increasing nutrient and water capture whilst reducing losses, root traits affecting soil exploration such as deep rooting and absorption are promising targets that are of particular importance for catch cover crops. Nitrate is a highly soluble form of N that leaches into the deeper soil layers in climates with high precipitation, and therefore, increasing effective nitrate interception and foraging at depth by roots is paramount (Kristensen & Thorup‐Kristensen, 2004; Trachsel et al., 2013). A linear relationship has been shown in the field between root density and 15N uptake at different depths as evaluated across three cover crop species: ryegrass, winter rye and fodder radish (Kristensen & Thorup‐Kristensen, 2004). Deep rooted perennial catch cover crops, tall fescue, chicory and lucerne, were shown to increase soil nutrient bioavailability in the topsoil with subsequent yield improvements of short‐season cereals grown in the plots (Han et al., 2021). Uplift of N by deep‐rooted cover crops is likely to be advantageous even if the cover crop root system is more extensive than the subsequent cash crop and able to forage further soil zones. Phosphate in comparison is highly immobile and is most abundant in the topsoil as plant available orthophosphate forms insoluble complexes with soil. Topsoil foraging by roots is an effective strategy for uptake of phosphate with greater growth and P accumulation in shallow‐rooted genotypes compared to deeper‐rooted genotypes (Ho et al., 2005; B. Sun et al., 2018; J. Zhu, et al., 2005). However, effectiveness of having a deep only or shallow only root system is specific to the environment and management strategy and incurs tradeoffs in multiple resource acquisition (Ho et al., 2005). In parallel with effective root exploration, roots also need to absorb nutrients from their surroundings. Heritable variation has been shown for specific nutrient uptake rates within species indicating that evaluation and genetic improvement of uptake will likely improve plant performance and yield while reducing fertiliser losses (Griffiths et al., 2021; Pace & McClure, 1986). Root evaluation during seedling establishment is also important as seedling vigour has been correlated with yield and quality in cash crops (Louvieaux et al., 2020; Thomas et al., 2016). For cover crops that overwinter, research programmes that focus on improving seedling vigour will be important as early capture of nutrients in the fallow period will benefit growth and development of the plant later in the season with fewer nutrient losses. Future cover crop research should focus on optimisation of the spatial distribution of roots in soil and root development processes that have implications on resource capture.
Another strategy to increase soil N is the use of legumes. Leguminous cover crops, such as pea, vetch and alfalfa, are specialists in facilitating biological N fixation with a symbiotic relationship with rhizobia (Table 1). Leguminous cover crops are often referred to as a 'green manure' fixing atmospheric N into plant usable forms at rates ranging between 30 and 280 kg ha−1 depending on species and conditions (Brady & Weil, 2002). Legumes can be used to improve N supply to succeeding cash crops, or they can be intercropped with a cash crop providing N fixation during growth. The use of legumes compared to catch cover crops is often considered situational as the effectiveness of legumes for N uptake falls with N supply. Therefore, reducing N losses with catch cover crops would be more effective than N fixation by a legume (Torbert et al., 1996; White et al., 2017). Legumes can also be effectively grown as part of a mixture with non‐legumes for N capture whilst adding more N to the system with fixation (Wittwer et al., 2017). Cover crop mixtures can be self‐regulating with a negative correlation between growth of legume and nonlegume species, and so, greater legume growth occurs in low N environments where biological N fixation is advantageous (De Notaris et al., 2021). Optimisation of nodulation traits including count, mass, morphology, and longevity are important targets for improving legume N fixation irrespective of high local nitrate presence (Herridge & Rose, 2000; Roy et al., 2020). For phosphorus, plant roots can release compounds into the soil environment and influence nutrient cycling with carboxylates promoting P mobilisation (Hinsinger et al., 2011). Plants in the Proteaceae family including white lupin (Lupinus albus) have highly specialised root systems that respond to P deficient soils by forming root clusters with large surface area for exuding nutrient‐solubilizing compounds (Shane & Lambers, 2005). In a meta‐analysis of multiple field experiments, services provided by cover crops were found to provide a measurable enhancement to main crop yield and P uptake with a greater benefit found in low available P environments (Hallama et al., 2019). Genetic improvement in cover crop N fixation rate and efficiency as well as P solubilisation could eventually displace chemical fertiliser application needs.
2.3. Maintenance of the rhizosphere and building soil organic matter content
By modifying the biotic and abiotic properties of the soil in which they grow, cover crops create soil legacies that can affect the ability of future plants to grow in that particular soil and interact with other organisms (Barel et al., 2018; Pineda et al., 2020). This phenomenon, referred to as plant‐soil feedback, is well known to ecologists and farmers. Negative plant‐soil feedbacks most often arise from nutrient depletion or the progressive build‐up of species‐specific soil pathogens in the rhizosphere of plants. These negative feedbacks determine the relative performance of plant species in a community and can affect crop yields when the same species is grown on the same soil for several years (van der Putten et al., 2013) as demonstrated with yield penalties found in constant corn and constant soybean fields (Seifert et al., 2017).
The effect of plants on the biotic and abiotic properties of the soil strongly depends on species as well as on plant traits (Baxendale et al., 2014; Cortois et al., 2016; Henneron, Cros et al., 2020). The quantity and quality of organic compounds that are exuded by plant roots vary among species and environmental conditions (Oburger & Jones, 2018; Williams & de Vries, 2020). For example, exudates collected from drought‐stressed plants showed increased soil respiration compared to exudates collected from well‐watered plants despite a lower exudation rate in stressed plants (de Vries et al., 2019). Recent evidence showed that interspecific differences in root exudation rates and composition can be partly explained by differences in root traits, with higher root exudation rates and rhizodeposition observed in productive species with a low root tissue density and a high root N concentration, such as legumes (Henneron, Cros, et al., 2020; L. Sun et al., 2021; Williams et al., 2021a). Root exudates have been shown to have a strong positive effect on soil carbon dynamics (de Vries et al., 2019; Henneron, Cros, et al., 2020; Henneron, Kardol, et al., 2020). Root exudation also plays a central role in creating soil microbial and chemical legacies (Delory et al., 2021). Plants mediate belowground biotic interactions and structure the rhizosphere microbiota through root exudation (Hu et al., 2018; Sasse et al., 2018; N.‐Q. Wang et al., 2021). For instance, while leguminous species can maintain associations with N2‐fixing bacteria and increase the abundance and diversity of arbuscular mycorrhizal fungi in the soil, non‐mycorrhizal species with a high glucosinolate content, such as brassicas, can have the opposite effect and decrease fungal diversity (Vukicevich et al., 2016).
Cover crop polycultures, where several cover crop species are planted at the same time, can provide multiple ecosystem services with increased benefits compared to monocultures. Long‐term biodiversity and ecosystem function experiments in ecology have repeatedly demonstrated the crucial role that plant species and functional group richness plays in enhancing ecosystem processes such as biomass production and biogeochemical cycles (Hector et al., 1999; Weisser et al., 2017). In comparison with low diversity mixtures, diverse plant communities often sustain a greater number of functions and services simultaneously, showing greater multifunctionality (Meyer et al., 2018). This positive relationship between biodiversity, ecosystem processes, and the provisioning of ecosystem services has motivated the development of cropping strategies aiming to increase multifunctionality in managed ecosystems by increasing biodiversity in both time and space (Barel et al., 2018; Finney & Kaye, 2017; Tiemann et al., 2015). In addition to increasing weed suppression and N retention (Finney & Kaye, 2017), cover crop polycultures are often more productive than their monoculture counterparts. Barel et al. (2018) found that two‐species cover crop polycultures overyielded in aboveground biomass (Raphanus sativus + Vicia sativa) or belowground biomass (Lolium perenne + Trifolium repens), but the legacy effect of the cover crop polyculture on the productivity of the following crop was dependent on the mixture of species used as a cover crop. Increased species complementarity has been one of the main causes cited to explain the positive effects of biodiversity on ecosystem functioning (Barry et al., 2019; Cardinale et al., 2007; Loreau & Hector, 2001). Species complementarity, however, encompasses several mechanisms that are likely to operate simultaneously in the field, such as resource partitioning, abiotic facilitation (e.g., microclimate amelioration and N facilitation by legumes), and biotic feedback (Barry et al., 2019). Although all these mechanisms may play a role in increasing complementarity between species grown in polycultures (Barry et al., 2019; Postma & Lynch, 2012), their relative contributions in determining positive biodiversity‐ecosystem functioning relationships in agroecosystems remain elusive.
As with root exudation, both the quantity and quality of the cover crop litter, which greatly vary among species, influence the biotic and abiotic properties of the soil, litter decomposition, and the performance of the subsequent crop (Barel et al., 2019; Haramoto & Gallandt, 2004). In a field experiment, Barel et al. (2018) found that the productivity of subsequent crops can be stimulated by winter cover crops with a high biomass production and a high shoot N concentration, such as the legumes T. repens and V. sativa. The use of cover crops in rotations can positively affect the soil microbiome by enhancing microbial biomass and activity (Barel et al., 2019; McDaniel et al., 2014; Tiemann et al., 2015), which can then contribute to the formation and greater stability of soil aggregates (Tiemann et al., 2015). Cover crop litter inputs in the soil modulate the structure and diversity of soil microbial communities. Litter decomposition rates by saprotrophic microbes also depend on plant litter composition as lignin and dry matter content have been shown to be negatively correlated to decomposability (Barel et al., 2019; Freschet et al., 2012; Vukicevich et al., 2016). Compounds produced during degradation of cover crop residues can also help mitigate weed proliferation in the field. For instance, glucosinolate hydrolysis products, such as isothiocyanates that are released by decomposing brassicas, have allelopathic properties and suppress weed populations by inhibiting or delaying seed germination, affecting plant establishment, and/or reducing plant growth (Haramoto & Gallandt, 2004). However, the non‐target impacts of biofumigation on biodiversity in the field are not clear (Henderson et al., 2009). Decomposition and mineralisation of cover crop residues in soils can increase soil organic matter content and improve soil fertility for the next crop (Barel et al., 2019). Although a decrease or a lack of change in soil organic matter content after cover crop cultivation has been reported in the short term (Barel et al., 2018), increasing agricultural crop diversity through the inclusion of cover crops in rotations can be an efficient way to increase soil organic carbon stocks (McDaniel et al., 2014; Tiemann et al., 2015). Cover crop mixtures were shown to improve soil health after 4 years in a continuous corn system, independent of soil tillage practiced (Nunes et al., 2018). Utilisation of cover crops meant that there was a longer period of time with living plants and roots in the agroecosystem which contributed to an increase in soil organic matter content and quality. Soil organic matter content has been ignored in recent years as high levels of fertiliser and irrigation practices are used to increase and maintain yields. Formation of higher soil organic matter content soils can offset this dependency on inputs for more sustainable agriculture. A diversified cropping system incorporating cover crops and polyculture offers soil health benefits and more sustainable crop production options for farmers.
3. PLANT SPECIES AND FUNCTIONAL GROUPS CURRENTLY IN USE AS COVER CROPS
Cover crop species at present span across the grasses, brassicas and legume families, all of which provide soil erosion protection compared to fallow land and each providing additional ecosystem functions (Table 1). In a single‐year harvest of multiple cover crop species, the phenotypic traits measured were found to cluster by plant family, with brassicas and legumes more closely associated in function compared to the grasses (Figure 2A). Details regarding our cover crop field study including images, data, and statistical analysis scripts are available at 10.5281/zenodo.5039308. Shoot biomass density, root mass fraction, root length and biomass density in topsoil, weed severity score, and shoot count were the greatest contributing traits to the first principal component explaining 46% of the variation. Total biomass and root biomass density in topsoil were the main contributing traits to the second principal component explaining 20% of the variation. For the root traits, the grasses had an overall higher root biomass density in the topsoil, a larger specific root length, and deeper maximum root length, which are ideal for preventing soil erosion and high nutrient capture (Figure 2B–D). Legumes, being the only family that can form nodules, provided a unique mechanism of action for N capture via N fixation. The grasses overall had the greatest total biomass production with a higher shoot biomass density, but the higher shoot biomass density did not translate into effective weed suppression (Figure 2E,F). The legumes and brassicas were the most effective at weed suppression (Figure 2F,G). High canopy cover in legumes may result in weed suppression, while the mechanism of weed suppression in brassicas is likely root exudation and biofumigation. Cover crops available at present are functionally diverse and vary in the eco‐agrosystem services that they provide. This diversity will affect the utilisation of a particular cover crop variety and choice of mixtures for multifunctionality.
Figure 2.
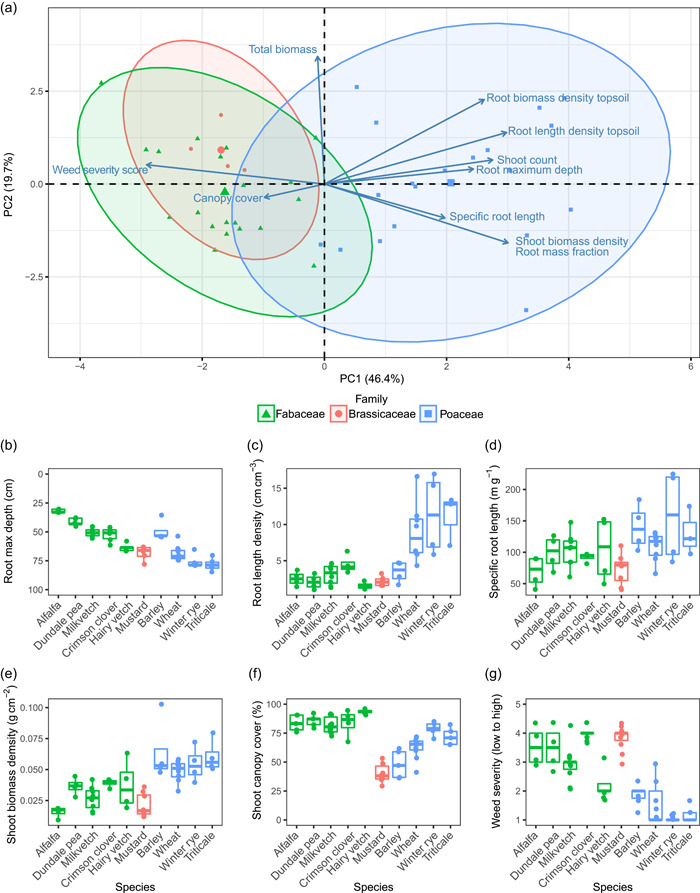
Phenotypic trait evaluation of cover crop species grown in a single field trial. (A) Principal component analysis of plant phenotypic traits clustered by family with root mass fraction and weed severity scores the greatest contributors to PC1 and total biomass having the greatest contribution to PC2. (B–G) Boxplots showing individual phenotypic trait scores per cover crop species. Alfalfa (Medicago sativa), dundale pea (Trifolium incarnatum), milkvetch (Astragalus spp.), crimson clover (Pisum sativum), hairy vetch (Vicia villosa), mustard (Brassica juncea), barley (Hordeum vulgare), wheat (Triticum aestivum), winter rye (Secale cereale) and triticale (×Triticosecale) [Color figure can be viewed at wileyonlinelibrary.com]
4. OPPORTUNITY FOR IMPROVEMENT OF COVER CROP ROOT TRAITS
Unlike major cash crops, which have passed through significant genetic bottlenecks during millennia of breeding, most species used as cover crops are untouched. As a result, the heritable genetic variance lost during domestication of cash crops remains available to most cover crop breeders. Targeted improvement in root resource capture, soil structure remediation, rhizosphere formation, biological N fixation, and carbon sequestration are highly desirable (Figure 3). However, this diversity also complicates traditional breeding efforts—not only are the ecosystem service targets of selection variably affected by cover crop species, environments, and cropping systems, but the genetic and physiological mechanisms that determine these traits often differ dramatically and non‐linearly across environments. Despite the great potential for cover crop improvement, selection for traits that consistently enhance ecosystem services represents a fundamental challenge to breeders. Development of phenotyping and genetic tools will be integral to characterising diverse root functional strategies of cover crops and facilitating genetic improvement.
Figure 3.
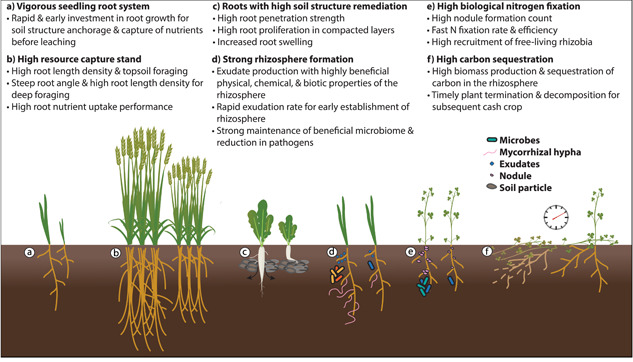
Root system ideotypes for cover crop species that provide greater ecosystem function [Color figure can be viewed at wileyonlinelibrary.com]
4.1. Phenotyping approaches for characterising cover crop root traits
Evaluation of root traits is important for assessing root function and selection of improved resource capture and abiotic stress tolerant varieties. However, root phenotyping is technically challenging and time consuming. In recent years image‐based root phenotyping approaches have become more widely utilised (Atkinson et al., 2019). Root phenotyping approaches have been primarily developed and utilised in phenotyping roots of model plant species, such as Arabidopsis thaliana, and cash crop species, such as maize, wheat and rice. These methods can be directly applied to cover crops with further development warranted for measuring ecosystem service performance. Phenotyping approaches vary widely in the realism of the growth system, infrastructure requirements, and information content of the extractable measurements (Topp et al., 2016). At present, there is no perfect system for evaluation of root system architecture, with each method having their own trade‐offs.
Methods of root evaluation range from high‐precision benchtop measurements to coarse field‐scale techniques. As roots are hidden in soil, much of the fundamental root research uses plants grown in clear agar, hydroponics, or seedling pouches. Growing in soil‐less media allows clear visualisation of roots from background, control of environment for treatment evaluation, and measurement of nutrient and root respiration fluxes with methods that are often high‐throughput (Paez‐Garcia et al., 2015). In contrast, evaluation of plants grown in soil is more representative but comes with the cost of often slower, coarser, and more destructive root analysis techniques (J. Zhu et al., 2011). Pot and tall mesocosm studies often require a root washing step to remove soil before root imaging, but this processing can destroy fine roots and affect the root system architecture. Nevertheless, these studies have been instrumental for evaluating rooting depth, root branching, resource capture and abiotic stress tolerance (Gao & Lynch, 2016; Guo & York, 2019; Zhan & Lynch, 2015). Root evaluation in the field is the most challenging with current approaches being mostly destructive excavation analyses that provide limited viewpoints of the root system. One field approach for root evaluation in the topsoil is called 'shovelomics' where a soil monolith is collected, washed, and imaged with crown root system architecture being discernable if strongly lignified (Das et al., 2015; Seethepalli et al., 2020; Trachsel et al., 2011), or the root length parameters determined without spatial orientation using a flatbed scanner (Seethepalli et al., 2021). For root evaluation at depth, soil coring is a widely used approach with root quantification either after a root washing and flatbed scanning or by a soil‐break method in combination with fluorescence spectroscopy imaging (Wasson et al., 2016). The gold standard approach to measure root distribution at depth in the field is trench excavation. Yet, the highly destructive and labour‐intensive nature prohibits wide utility (Böhm 1979; van Noordwijk et al., 2001). Proxies have also been used to predict root length. For example, shoot manganese concentration positively correlated with carboxylate exudation in the rhizosheath and root size traits in chickpea (J. Pang et al., 2018).
Non‐destructive root analyses are particularly valuable as the root system architecture and root processes are intact, and the same plant can be analysed across time. While most non‐destructive root phenotyping approaches use plants grown in artificial environments (Nagel et al., 2012), soil‐grown methods include rhizotrons and field minirhizotrons also provide 2D snapshots of the roots in contact with a clear wall (Arnaud et al., 2019). Specialised equipment is required to extract the 3D root system architecture deep in soil without disturbing root structure. Technologies such as X‐ray computed tomography (CT), magnetic resonance imaging (MRI) and positron emission tomography (PET) are proving to be useful for non‐destructive root 4D view of the rhizosphere (Duncan et al., 2021; Helliwell et al., 2017; Pflugfelder et al., 2017; N.‐Q. Wang et al., 2014; Zhou et al., 2021). Combining multiple imaging modalities, such as PET‐CT, is an emerging direction that can observe both structural and biological function such as nutrient flow with nutrient uptake to understand more complex rooting behaviour (Garbout et al., 2012). Scaling up these high‐precision approaches to be high‐throughput is a current challenge with complex segmentation analysis and a tradeoff in scan resolution with pot size and scan time. Common to all root imaging techniques described, image analysis is often the limiting factor. Deep learning approaches are now allowing faster and more accurate plant image analysis (Han et al., 2021; Smith et al., 2021; Soltaninejad et al., 2020). Promising applications that can be scaled up in the field include electrical resistance tomography, electromagnetic inductance and ground penetrating radar; however, these methods require species‐dependent optimisation as many factors affect root biomass estimations including soil texture, soil water content, and organic matter (X. Liu et al., 2018; Weigand & Kemna, 2017).
Advancements in root phenotyping approaches show promise in detangling individual contributions in polyculture systems. Root characterisation of polycultures can help in understanding the mechanisms of species complementarity and in designing effective species mixtures under various environmental conditions. Due to the difficulty in visually separating roots of different species and obtaining species‐specific data on root distribution in the soil, determining the extent to which species grown in mixtures partition belowground resources in time and space has always been a very challenging task in ecology and agriculture (Rewald et al., 2012). In recent years, direct tests of spatial resource partitioning between species have been made possible due to technological breakthroughs allowing the quantification of species relative abundances in mixed root samples. Both Fourier‐transform mid‐infrared attenuated total reflection spectroscopy (Meinen & Rauber, 2015) and the amplification of species‐specific DNA sequences using reverse transcription polymerase chain reaction (RT‐PCR) (Haling et al., 2011; Heuermann et al., 2019; Mommer et al., 2008, 2010) have been successfully used for this purpose. However, both methods have a relatively low throughput and require a labour‐intensive calibration that prevents their use in species‐rich mixtures (Rewald et al., 2012). Recently, a new high‐throughput next generation sequencing‐based method was developed to quantify species proportions in mixed root samples (Wagemaker et al., 2021). This method, referred to as multispecies genotyping by sequencing (msGBS), proved to be as accurate as the PCR‐based method of (Mommer et al., 2008). It is also more sensitive and less labour‐intensive. In addition, it does not require the development of species‐specific DNA primers, which makes it possible to analyse root samples collected from species‐rich polycultures and measure traits such as rooting depth for each species individually (in‘t Zandt et al., 2020; Wagemaker et al., 2021). Root phenotyping in polycultures can also be facilitated using crop lines that were genetically modified to express a green or red fluorescent protein (Faget, Nagel, et al., 2013). For each transformed species, this approach allows the visual detection of roots growing in rhizoboxes or along minirhizotron tubes (Faget et al., 2009; Faget, Blossfeld, et al., 2013), which makes it possible to non‐destructively measure morphological and architectural root traits as well as root proliferation in different soil zones (Weidlich et al., 2018).
In recent years, advances in root imaging approaches have shown great promise for evaluation of root system architecture. Evaluation of rhizosphere processes such as nutrient cycling, root exudation, and microbiome changes are also important for improving ecosystem service performance. Stable isotope labelling can be used with mass spectrometry to trace C and N pools in plant or soil samples. Changes in the 13C/12C ratio in root‐adhering soil can be used to quantify root exudation rates for plants labelled with 13CO2 (Guyonnet et al., 2018; R. Pang et al., 2021). In recent years, nanoscale secondary ion mass spectrometry (NanoSIMS) has become popular as it enables simultaneous imaging and isotopic discrimination of stable isotopes at a subcellular resolution (Kilburn et al., 2010). For quantifying nutrient fluxes by plants and the respiration cost of roots, recent experimental protocols have been scaled up to phenotype large mapping populations (Griffiths et al., 2021; Guo et al., 2021). At a broader scale, continuous in situ soil nitrate sensors are showing promise to track the fate of fertiliser across the year (Y. Zhu et al., 2021). For root exudation and P solubilisation estimation, carboxylates and acid phosphatase activity can be estimated from collected roots and rhizosheath (soil that tightly adheres to root, Shen et al., 2003; Wen et al., 2019). Root exudation profiling approaches from soil grown plants currently have low reproducibility, and therefore, artificial systems such as hydroponic culture accumulate sufficient quantities of exudate for isolation and identification by liquid chromatography‐mass spectrometry (LC‐MS), despite the exudation profile being less representative compared to soil (Vives‐Peris et al., 2020; Williams et al., 2021b; Zhalnina et al., 2018). Root colonisation by AMF can be quantified by root intersection method on roots stained with Trypan blue (Freschet et al., 2021; McGonigle et al., 1990; Walker, 2005). For identification of beneficial soil microbes, 16S and ITS rRNA gene amplicon sequencing analysis can be used for bacteria, archaea, and fungi in the root microbiome (George et al., 2019; McPherson et al., 2018). Isolated root exudates and microbes from field soil can then be used in more controlled experiments to functionally characterise modes of action on the plant and rhizosphere (Hao, Zhang, et al., 2021; Macias‐Benitez et al., 2020).
A promising research direction is in developing a 'controlled field' system that bridges between plants in single pots and the field. Pot experiments are inherently limited by the container size; they have pot‐bound roots and lack neighbouring plant competition as would be found in the field. In contrast, root phenotyping approaches in the field are challenging, lack precision, and lack control of environment parameters. A modern mesocosm system has been fitted with moisture, gas, and temperature sensors allowing for daily tracking of environmental fluxes and measurement of unrestricted 3D root system architecture (Dowd et al., 2021). Advancing this concept with the development of a larger 'controlled field' system will allow for crop stand evaluation of root and ecosystem service performance, such as resource capture, in a more representative and high‐precision manner. Incorporating these root phenotyping systems with functional traits and ecosystem service measurements will be important for evaluating and improving cover crop root systems. Phenotyping of stand root traits, such as root length density and biomass per volume soil, is more representative of field scale ecosystem service performance than working with single plants in isolation. Coupling root system architecture analyses of unrestricted roots with 15N and 2H dual‐labelling approaches or nitrate sensors will also provide functional insights into how stand establishment and root phenotypes affect short‐term dynamics of water and N uptake (Chen et al., 2021; Y. Zhu et al., 2021).
4.2. Leveraging genetic variation and tools to improve cover crop root traits
The underlying genetic variation behind key traits is fundamental to generating improved cultivars. Genetic bottlenecks have regularly occurred during domestication of major crop species, such as maize, wheat, rice and soybean (Caicedo et al., 2007; Eyre‐Walker et al., 1998; Haudry et al., 2007; Hyten et al., 2006). Efforts have been made to introgress near‐ and distant‐ancestral material back into modern crops to reintroduce genetic diversity that has been lost (Reynolds et al., 2009; X. Wang et al., 2019; Yang et al., 2019). Cover crops have undergone minimal domestication and genetic selection compared to cash crops and represent an opportunity to incorporate more genetic variation from the outset. Collections of natural accessions and wild relatives are often available and can serve as reservoirs of genetic diversity during the ongoing domestication of cover crop species. Multivariate analysis reports that modern pea cultivars have a close genetic relationship to each other in contrast to the diversity detected in accession collections (Smýkal et al., 2012). Pea species can be crossed with wild varieties to enrich the genetic pool to prevent further loss of genetic diversity; cowpea has over 200 wild relatives and wild Pisum fulvum, Pisum sativum ssp. elatius, Pisum sativum ssp. sativum and Pisum abyssinicum could be introgressed with domesticated pea, P. sativum, to increase genetic diversity (Tani et al., 2017). Thlaspi arvense (pennycress) has extensive resources as diverse germplasms have been collected worldwide and over 500 natural variants have been sequenced (Frels et al., 2019). Vast genetic variation can be maintained in Triticale (× Triticosecale), a hybrid of wheat and rye, by crossing together cultivars of wheat and rye as well as crossing to diploid, tetraploid or hexaploid wheat varieties (Ayalew et al., 2018; Baker et al., 2020; Oettler et al., 1991). Selection for improved traits while maintaining genomic diversity is crucial for long‐term success of cultivars to preserve alleles that may confer adaptability under shifting environmental conditions or resource availability. New phenotyping methods, an increased attention to root structure and biomass, and large sources of genetic diversity in cover crops can be leveraged to create elite cultivars with desirable above and belowground traits (Figure 3).
Identifying and understanding the molecular basis of phenotypic diversity of root traits is advantageous for breeding cover crop cultivars with improved ecosystem services and must precede precise trait introgression or engineering efforts through synthetic biology approaches. Modern breeding programmes often incorporate molecular or genomics‐assisted breeding to efficiently generate improved varieties. Genetic resources for cover crop species are not as extensive as those for major crops but many species now have reference genomes, transcriptomes, and genetic tools—including single‐nucleotide polymorphism (SNP) maps and diversity panels to perform genome‐wide association studies (GWAS) and quantitative trait locus (QTL) analysis—to conduct functional genomic analysis (Ayalew et al., 2018; Boukar et al., 2016; Frels et al., 2019; Jones et al., 2020; Smýkal et al., 2012). Literature from related species can be exploited to direct breeding or targeted modification by identifying candidate genes. For example, greater than 70% of the predicted peptides of the pennycress genome have more than 80% sequence similarity to those of A. thaliana, and mutations in homologues result in comparable phenotypes (Chopra et al., 2018; Dorn et al., 2015). Triticale has a similar advantage as functional studies from wheat and rye are directly applicable, but phenotypes may be less predictable due to the multiploidy and hybrid nature of its genome (Mergoum et al., 2009; Oettler et al., 1991).
Improved agronomic traits are often determined by SNPs or a few base changes in a single gene (Doebley et al., 2006; J. Li et al., 2017; Mishra et al., 2020). Mutagens can introduce genetic diversity in species where genomic resources are limited or gene editing technology is not a viable tool. Random mutagenesis with radiation or chemical mutagens has been used since the 1920s to perform forward genetic screens for mutants with improved traits and has resulted in hundreds of released crop varieties (Holme et al., 2019). Reverse genetics with a mutagenized population is now possible using targeting induced local lesions in genomes (TILLING) to identify mutations in any gene of interest (Holme et al., 2019; Jankowicz‐Cieslak et al., 2017; Kurowska et al., 2011). TILLING is not limited by species or genome size making it a promising tool to identify novel alleles in cover crops. An ethyl methanesulfonate (EMS) mutant library has already been generated for pennycress and will accelerate identification of genetic alleles to improve root architecture or function (Marks et al., 2021). The integration of 'speed breeding' approaches that use prolonged photoperiods and early harvesting of seed to reduce generation time in cover crops species would accelerate the generation of inbred lines (Hickey et al., 2019; Wasson et al., 2016).
Gene editing technologies can accelerate trait improvement through precise editing of genes to introduce novel or proven alleles. Clustered regularly interspaced short palindromic repeats (CRISPR) technology allows for the simultaneous targeting of unique genes to multiplex alleles in a single generation to alter independent traits and circumvents linkage constraints encountered in crosses (Cong et al., 2013; Jinek et al., 2012; Lowder et al., 2015; Ma et al., 2015; Xing et al., 2014). CRISPR has most widely been used to edit coding regions to generate null alleles, however, editing of cis‐regulatory elements is also advantageous and often yields subtle phenotypic changes desired by breeders with fewer pleiotropic effects (Rodríguez‐Leal et al., 2017; Vats et al., 2019). Powerful new technologies, such as base editing and prime editing, precisely target specific residues of the defined sequence to introduce a SNP or a specified edit at single‐base resolution (Mishra et al., 2020). Prime editing uses a template to directly integrate a desired genetic sequence into the specified genomic target. Prime editing is being optimised in plants and has the potential to generate insertions, deletions, and all possible base substitutions to engineer genes (Anzalone et al., 2019; Hao et al., 2021; H. Li et al., 2020; Lin et al., 2021; Tang et al., 2020). The power of functional genomics combined with gene editing technologies can be harnessed to rapidly domesticate cover crops and improve root traits, particularly in species with limited natural genetic diversity. Applying the capabilities of gene editing tools is primarily limited by the capacity to transform the target species. Pennycress is well‐positioned for direct genetic modification due to its ease of transformation, diploid genome, low genetic redundancy and self‐fertility allowing for propagation of true breeding lines. Transformation of pennycress via a floral dip method has already been optimised (McGinn et al., 2019) and lines expressing transgenes or harbouring novel gene knockouts generated with CRISPR have been used to manipulate seed oil profiles (Jarvis et al., 2021). Together, this supports that pennycress is malleable and well‐suited for targeted genetic modifications to enhance and stack desirable traits. Transformation of crimson clover, T. pratense, and white cover, T. repens, has been accomplished through callus induction to regenerate plants with disrupted flavonoid production to investigate the role of flavonoids in nodule formation (Dinkins et al., 2021), but is most successful in lines optimised for regeneration in tissue culture. Trifolium is an outcrosser meaning it cannot self‐fertilise, and therefore, transgenic lines must be clonally propagated to maintain the transgene or gene edits. Alternatively, mating with a line carrying a self‐compatible locus increases successful mating between the progeny to generate homozygous lines (Dinkins et al., 2021; Riday & Krohn, 2010), but this method limits introgression of diverse genetic backgrounds. Preliminary studies in hairy vetch demonstrate that transformation may be possible but have yet to produce a stable transgenic line (Nguyen & Searle, 2022). Transformation in monocot systems has become routine in major cereal crops but is at its infancy in most cover crop varieties. Transformation protocols for oat (Dattgonde et al., 2019; Gasparis et al., 2008), buckwheat (Kojima et al., 2000), and wheat and triticale (Hensel et al., 2009; Nadolska‐Orczyk et al., 2005) are available; all require tissue culture and plant regeneration with varying efficiencies. Thus, advancing transformation capabilities of cover crop species will reduce barriers to use gene editing technology for trait improvement.
Information about genes affecting root function can be leveraged to modify cover crops and improve root traits. It was recently demonstrated that roots of ethylene‐insensitive mutants in rice and Arabidopsis continued to elongate in compacted soil unlike those of wild‐type (Pandey et al., 2021) suggesting that subtle alleles of genes involved in ethylene signalling may be promising targets to improve rooting depth in compacted soils. Homologues of DEEPER ROOTING 1 (DRO1) are found across diverse phyla and are prime candidates to mine for, or create alleles in, cis‐regulatory and genic regions that may confer desirable changes in root depth, root angle, and root biomass at depth (Arai‐Sanoh et al., 2014; Guseman et al., 2017; Uga et al., 2013). With the expansion of genetic tools and resources for cover crops, identifying the underlying genetics controlling root traits and alleles that confer superior root traits are possible and will facilitate rapid improvement of cover crop root function.
4.3. Potential for 'phenomic selection' to accelerate gains in ecosystem service
With technological advancement and decreasing costs in high‐throughput genotyping tools, genomic selection is possible for many species. Diploid annual cover crops such as pennycress (T. arvense) are particularly amenable to available genetic improvement (McGinn et al., 2019). However, there are many cover crop species that are outcrossers, have high levels of heterozygosity, and are polyploid, in which development and implementation of genomic tools comes at high cost. For such cover crops with large, complex genomes, an alternative plant selection methodology called 'phenomic selection' is promising (Rincent et al., 2018). Phenomic selection can cheaply screen many individuals and make selections with no pre‐existing genetic data, which could potentially reduce or eliminate the need for genotyping in some cases (Rincent et al., 2018). Instead of relying on genetic variances, phenomic selection uses an indirect phenotypic measurement as a selection index for other traits of interest. Phenotypic variables can be used directly to replace genetic marker information in traditional selection methodology and thus can be applied cheaply and widely to many species (Rincent et al., 2018). As an example, near‐infrared reflectance spectroscopy has been successfully used to predict grain yield and heading date (Rincent et al., 2018). This methodology appears to be a reliable and consistent method that can help breeders in ranking and selecting varieties in breeding programmes (Lane et al., 2020). For widespread adoption of cover crops by farmers, an array of species and varieties are needed that are optimised for different climates, soil types, cropping systems, and ecosystem services (Thorup‐Kristensen et al., 2003). A phenomic selection methodology could help accelerate cover crop breeding efforts by domesticating locally adapted species without the need for any genotypic tools. Utilising native species as cover crops would be beneficial in that they can be locally adapted, ecologically diverse, and restore the habitat. Currently, only spectra from near‐infrared reflectance spectroscopy have been used as a phenomic selection model input. Phenotypic values from other root phenotyping methods may be utilised to predict plant performance traits. Development and implementation of a phenomic selection pipeline for ecosystem service performance could provide a framework for rapid and wide improvement of cover crop species. In addition, phenomic selection could facilitate simultaneous selection for multiple species, individually or in mixtures.
5. CONCLUSION AND FUTURE PROSPECTS
For agriculture sustainability, protecting soil health and reducing our dependence on chemical fertilisers is paramount. To attain this goal, the ecosystem services provided by cover crops; soil structural remediation, capture of soil resources, maintenance of the rhizosphere, and building soil organic matter content have the potential to provide an immediate and high impact to the farmer at a low cost. Understanding mechanisms of action behind ecosystem services and how plants coordinate their diverse root functional traits will be important for improving ecosystem service performance (Wen et al., 2019). Cover crop varieties available today have generally had very minimal genetic improvement from wild species and may be amenable to rapid genetic gain. With recent advancements in genetic tools, the domestication process can be accelerated by the stacking of multiple traits (Chopra et al., 2020). In addition, advancement in phenotyping approaches allow the simultaneous selection of root and shoot traits and devising cover crop mixes that provide multifunctional ecosystem services and enhance functional diversity in the field. Most cover crops, at present, are investment strategies with yield benefits to the cash crops realised after several years. Encouragingly, some ecosystem services provided by cover crops show immediate tractable benefits such as a strong reduction in soil erosion, N leaching, and weed suppression (Osipitan et al., 2019; Thorup‐Kristensen et al., 2012). Widespread adoption of cover cropping systems is unlikely to occur without short‐term economic gains, and therefore, 'cash cover crops' such as pennycress could provide the required financial incentive. In addition, government‐ and industry‐level support to cut greenhouse gas emissions from agriculture may stimulate adoption by offering payments to farmers embracing conservation practices (Reuters, 2021; The Wall Street Journal, 2021).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Christopher N. Topp and Marcus Griffiths conceived the manuscript topic, Marcus Griffiths generated the figures and table, and all authors contributed to the writing and revised the manuscript.
ACKNOWLEDGMENTS
The authors would like to thank Matthew J Rubin, Elisa Morales, Emelyn Piotter, Shalya Gunn, Keith Duncan, Tiffany Hopkins, Toni Johnson and Eric Byas Jr for sampling assistance of the cover crop field data. This study was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomic Science Programme grant no. DE‐SC0021286 to Christopher N. Topp and Dmitri A. Nusinow.
Griffiths, M. , Delory, B.M. , Jawahir, V. , Wong, K.M. , Bagnall, G.C. , Dowd, T.G. et al. (2022) Optimization of root traits to provide enhanced ecosystem services in agricultural systems: a focus on cover crops. Plant, Cell & Environment, 45, 751–770. 10.1111/pce.14247
Benjamin M. Delory and Vanessica Jawahir contributed equally to this study
DATA AVAILABILITY STATEMENT
Images, data, and statistical analysis scripts available at 10.5281/zenodo.5039308
REFERENCES
- Abdalla, M. , Hastings, A. , Cheng, K. , Yue, Q. , Chadwick, D. & Espenberg, M. et al. (2019) A critical review of the impacts of cover crops on nitrogen leaching, net greenhouse gas balance and crop productivity. Global Change Biology, 25, 2530–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamopoulos, T. & Restuccia, D. (2021) Geography and Agricultural Productivity: cross‐Country Evidence from Micro Plot‐Level Data. National Bureau of Economic Research. Working Paper No. 24532.
- Anzalone, A.V. , Randolph, P.B. , Davis, J.R. , Sousa, A.A. , Koblan, L.W. , Levy, J.M. et al. (2019) Search‐and‐replace genome editing without double‐strand breaks or donor DNA. Nature, 576, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai‐Sanoh, Y. , Takai, T. , Yoshinaga, S. , Nakano, H. , Kojima, M. , Sakakibara, H. et al. (2014) Deep rooting conferred by DEEPER ROOTING 1 enhances rice yield in paddy fields. Scientific Reports, 4, 5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravena, J.E. , Berli, M. , Ruiz, S. , Suárez, F. , Ghezzehei, T.A. & Tyler, S.W. (2014) Quantifying coupled deformation and water flow in the rhizosphere using X‐ray microtomography and numerical simulations. Plant and Soil, 376, 95–110. [Google Scholar]
- Arnaud, M. , Baird, A.J. , Morris, P.J. , Harris, A. & Huck, J.J. (2019) EnRoot: a narrow‐diameter, inexpensive and partially 3D‐printable minirhizotron for imaging fine root production. Plant Methods, 15, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, J.A. , Pound, M.P. , Bennett, M.J. & Wells, D.M. (2019) Uncovering the hidden half of plants using new advances in root phenotyping. Current Opinion in Biotechnology, 55, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew, H. , Kumssa, T.T. , Butler, T.J. & Ma, X.‐F. (2018) Triticale improvement for forage and cover crop uses in the Southern Great Plains of the United States. Frontiers in Plant Science, 9, 1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, L. , Grewal, S. , Yang, C.‐Y. , Hubbart‐Edwards, S. , Scholefield, D. , Ashling, S. , et al. (2020) Exploiting the genome of Thinopyrum elongatum to expand the gene pool of hexaploid wheat. Theoretical and Applied Genetics, 133, 2213–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barel, J.M. , Kuyper, T.W. , de Boer, W. , Douma, J.C. & De Deyn, G.B. (2018) Legacy effects of diversity in space and time driven by winter cover crop biomass and nitrogen concentration. The Journal of Applied Ecology, 55, 299–310. [Google Scholar]
- Barel, J.M. , Kuyper, T.W. , Paul, J. , de Boer, W. , Cornelissen, J.H.C. & De Deyn, G.B. (2019) Winter cover crop legacy effects on litter decomposition act through litter quality and microbial community changes. The Journal of Applied Ecology, 56, 132–143. [Google Scholar]
- Barry, K.E. , Mommer, L. , van Ruijven, J. , Wirth, C. , Wright, A.J. , Bai, Y. et al. (2019) The future of complementarity: disentangling causes from consequences. Trends in Ecology & Evolution, 34, 167–180. [DOI] [PubMed] [Google Scholar]
- Baxendale, C. , Orwin, K.H. , Poly, F. , Pommier, T. & Bardgett, R.D. (2014) Are plant‐soil feedback responses explained by plant traits? The New Phytologist, 204, 408–423. [DOI] [PubMed] [Google Scholar]
- Böhm, W. (1979) Profile wall methods. In: methods of studying root systems. ecological studies (analysis and synthesis) 33. Berlin, Heidelberg: Springer. [Google Scholar]
- Bommarco, R. , Kleijn, D. & Potts, S.G. (2013) Ecological intensification: harnessing ecosystem services for food security. Trends in Ecology & Evolution, 28, 230–238. [DOI] [PubMed] [Google Scholar]
- Boukar, O. , Fatokun, C.A. , Huynh, B.L. , Roberts, P.A. & Close, T.J. (2016) Genomic tools in cowpea breeding programs: status and perspectives. Frontiers in Plant Science, 7, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, N.C. & Weil, R.R. (2002) The nature and properties of soils. Upper Saddle River, NJ: Prentice Hall. [Google Scholar]
- Bünemann, E.K. , Bongiorno, G. , Bai, Z. , Creamer, R.E. , De Deyn, G. & de Goede, R. (2018) Soil quality—a critical review. Soil Biology and Biochemistry, 120, 105–125. [Google Scholar]
- Burr‐Hersey, J.E. , Mooney, S.J. , Bengough, A.G. , Mairhofer, S. & Ritz, K. (2017) Developmental morphology of cover crop species exhibit contrasting behaviour to changes in soil bulk density, revealed by X‐ray computed tomography. PLoS One, 12, e0181872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A.L. , Williamson, S.H. , Hernandez, R.D. , Boyko, A. , Fledel‐Alon, A. , York, T.L. et al. (2007) Genome‐wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genetics, 3, 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale, B.J. , Wright, J.P. , Cadotte, M.W. , Carroll, I.T. , Hector, A. , Srivastava, D.S. et al. (2007) Impacts of plant diversity on biomass production increase through time because of species complementarity. Proceedings of the National Academy of Sciences of the United States of America, 104, 18123–18128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati, A. , Moradi, A.B. , Vetterlein, D. , Vontobel, P. , Lehmann, E. , Weller, U. et al. (2010) Dynamics of soil water content in the rhizosphere. Plant and Soil, 321, 163–176. [Google Scholar]
- Cassman, K.G. , Dobermann, A.R. & Walters, D.T. (2002) Agroecosystems, nitrogen‐use efficiency, and nitrogen management. Ambio, 31, 132–140. [DOI] [PubMed] [Google Scholar]
- Chen, G. , Dresbøll, D.B. & Thorup‐Kristensen, K. (2021) Dual labelling by 2H and 15N revealed differences in uptake potential by deep roots of chicory. Rhizosphere, 19, 100368. [Google Scholar]
- Chen, G. & Weil, R.R. (2011) Root growth and yield of maize as affected by soil compaction and cover crops. Soil and Tillage Research, 117, 17–27. [Google Scholar]
- Chopra, R. , Johnson, E.B. , Daniels, E. , McGinn, M. , Dorn, K.M. , Esfahanian, M. et al. (2018) Translational genomics using Arabidopsis as a model enables the characterization of pennycress genes through forward and reverse genetics. The Plant Journal, 96, 1093–1105. [DOI] [PubMed] [Google Scholar]
- Chopra, R. , Johnson, E.B. , Emenecker, R. , Cahoon, E.B. , Lyons, J. , Kliebenstein, D.J. et al. (2020) Identification and stacking of crucial traits required for the domestication of pennycress. Nature Food, 1, 84–91. [Google Scholar]
- Comerford, N.B. (2005) Soil factors affecting nutrient bioavailability. BassiriRad H. (Ed.) Nutrient acquisition by plants. Ecological studies (Analysis and Synthesis) Springer, Berlin: Heidelberg, Vol. 181. 10.1007/3-540-27675-0_1 [DOI]
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortois, R. , Schröder‐Georgi, T. , Weigelt, A. , van der Putten, W.H. & De Deyn, G.B. (2016) Plant‐soil feedbacks: role of plant functional group and plant traits. The Journal of Ecology, 104, 1608–1617. [Google Scholar]
- Das, A. , Schneider, H. , Burridge, J. , Ascanio, A.K.M. , Wojciechowski, T. , Topp, C.N. et al. (2015) Digital imaging of root traits (DIRT): a high‐throughput computing and collaboration platform for field‐based root phenomics. Plant Methods, 11, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dattgonde, N. , Tiwari, S. , Sapre, S. & Gontia‐Mishra, I. (2019) Genetic transformation of Oat mediated by is enhanced with sonication and vacuum infiltration. Iranian Journal of Biotechnology, 17, e1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Notaris, C. , Mortensen, E.Ø. , Sørensen, P. , Olesen, J.E. & Rasmussen, J. (2021) Cover crop mixtures including legumes can self‐regulate to optimize N2 fixation while reducing nitrate leaching. Agriculture, Ecosystems and Environment, 309, 107287. [Google Scholar]
- Delory, B.M. , Schempp, H. , Spachmann, S.M. , Störzer, L. , van Dam, N.M. , Temperton, V.M. et al. (2021) Soil chemical legacies trigger species‐specific and context‐dependent root responses in later arriving plants. Plant, Cell & Environment, 44, 1215–1230. [DOI] [PubMed] [Google Scholar]
- Dinkins, R.D. , Hancock, J. , Coe, B.L. , May, J.B. , Goodman, J.P. , Bass, W.T. et al. (2021) Isoflavone levels, nodulation and gene expression profiles of a CRISPR/Cas9 deletion mutant in the isoflavone synthase gene of red clover. Plant Cell Teports, 40, 517–528. [DOI] [PubMed] [Google Scholar]
- Dodds, W.K. (2006) Nutrients and the “dead zone”: the link between nutrient ratios and dissolved oxygen in the northern Gulf of Mexico. Frontiers in Ecology and the Environment, 4, 211–217. [Google Scholar]
- Doebley, J.F. , Gaut, B.S. & Smith, B.D. (2006) The molecular genetics of crop domestication. Cell, 127, 1309–1321. [DOI] [PubMed] [Google Scholar]
- Dorn, K.M. , Fankhauser, J.D. , Wyse, D.L. & Marks, M.D. (2015) A draft genome of field pennycress (Thlaspi arvense) provides tools for the domestication of a new winter biofuel crop. DNA Research, 22, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd, T. , McInturf, S. , Li, M. & Topp, C.N. (2021) Rated‐M for mesocosm: allowing the multimodal analysis of mature root systems in 3D. Emerging Topics in Life Sciences, 2, 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, K.E. , Czymmek, K.J. , Jiang, N. , Thies, A.C. & Topp, C.N. (2021) X‐ray microscopy enables multiscale high‐resolution 3D imaging of plant cells, tissues, and organs. Plant Physiology, 16, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle, C.A. , Thom, M.D. , Nemec, K.T. , Forcella, F. , Lundgren, J.G. , Gesch, R.W. et al. (2015) Using pennycress, camelina, and canola cash cover crops to provision pollinators. Industrial Crops and Products, 75, 20–25. [Google Scholar]
- Eyre‐Walker, A. , Gaut, R.L. , Hilton, H. , Feldman, D.L. & Gaut, B.S. (1998) Investigation of the bottleneck leading to the domestication of maize. Proceedings of the National Academy of Sciences of the United States of America, 95, 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faget, M. , Blossfeld, S. , von Gillhaussen, P. , Schurr, U. & Temperton, V.M. (2013) Disentangling who is who during rhizosphere acidification in root interactions: combining fluorescence with optode techniques. Frontiers in Plant Science, 4, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faget, M. , Herrera, J.M. , Stamp, P. , Aulinger‐Leipner, I. , Frossard, E. & Liedgens, M. (2009) The use of green fluorescent protein as a tool to identify roots in mixed plant stands. Functional Plant Biology, 36, 930–937. [DOI] [PubMed] [Google Scholar]
- Faget, M. , Nagel, K.A. , Walter, A. , Herrera, J.M. , Jahnke, S. , Schurr, U. et al. (2013) Root‐root interactions: extending our perspective to be more inclusive of the range of theories in ecology and agriculture using in‐vivo analyses. Annals of Botany, 112, 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO . (2021). Land statistics. Global, regional and country trends, 1990–2018. FAOSTAT Analytical Brief Series No. 15.
- Finney, D.M. & Kaye, J.P. (2017) Functional diversity in cover crop polycultures increases multifunctionality of an agricultural system. The Journal of Applied Ecology, 54, 509–517. [Google Scholar]
- Frels, K. , Chopra, R. , Dorn, K.M. , Wyse, D.L. , David Marks, M. & Anderson, J.A. (2019) Genetic diversity of field pennycress (Thlaspi arvense) reveals untapped variability and paths toward selection for domestication. Agronomy, 9, 302. [Google Scholar]
- Freschet, G.T. , Aerts, R. & Cornelissen, J.H.C. (2012) A plant economics spectrum of litter decomposability. Functional Ecology, 26, 56–65. [Google Scholar]
- Freschet, G.T. , Pagès, L.L. , Iversen, C.M. , Comas, L.H. , Rewald, B. & Mccormack, M.L. (2021) A starting guide to root ecology: strengthening ecological concepts and standardising root classification, sampling, processing and trait measurements. New Phytologist, 232, 973–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. & Lynch, J.P. (2016) Reduced crown root number improves water acquisition under water deficit stress in maize (Zea mays L.). Journal of Experimental Botany, 67, 4545–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbout, A. , Munkholm, L.J. , Hansen, S.B. , Petersen, B.M. , Munk, O.L. & Pajor, R. (2012) The use of PET/CT scanning technique for 3D visualization and quantification of real‐time soil/plant interactions. Plant and Soil, 352, 113–127. [Google Scholar]
- Gasparis, S. , Bregier, C. , Orczyk, W. & Nadolska‐Orczyk, A. (2008) Agrobacterium‐mediated transformation of oat (Avena sativa L.) cultivars via immature embryo and leaf explants. Plant Cell Reports, 27, 1721–1729. [DOI] [PubMed] [Google Scholar]
- George, P.B.L. , Creer, S. , Griffiths, R.I. , Emmett, B.A. , Robinson, D.A. & Jones, D.L. (2019) Primer and database choice affect fungal functional but not biological diversity findings in a national soil survey. Frontiers of Environmental Science & Engineering in China, 7, 173. [Google Scholar]
- Griffiths, M. , Roy, S. , Guo, H. , Seethepalli, A. , Huhman, D. , Ge, Y. et al. (2021) A multiple ion‐uptake phenotyping platform reveals shared mechanisms affecting nutrient uptake by roots. Plant Physiology, 185, 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H. , Ayalew, H. , Seethepalli, A. , Dhakal, K. , Griffiths, M. , Ma, X.‐F. et al. (2021) Functional phenomics and genetics of the root economics space in winter wheat using high‐throughput phenotyping of respiration and architecture. The New Phytologist, 232, 98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H. & York, L.M. (2019) Maize with fewer nodal roots allocates mass to more lateral and deep roots that improve nitrogen uptake and shoot growth. Journal of Experimental Botany, 70, 5299–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurr, G.M. , Lu, Z. , Zheng, X. , Xu, H. , Zhu, P. , Chen, G. et al. (2016) Multi‐country evidence that crop diversification promotes ecological intensification of agriculture. Nature Plants, 2, 16014. [DOI] [PubMed] [Google Scholar]
- Guseman, J.M. , Webb, K. , Srinivasan, C. & Dardick, C. (2017) DRO1 influences root system architecture in Arabidopsis and Prunus species. The Plant Journal, 89, 1093–1105. [DOI] [PubMed] [Google Scholar]
- Guyonnet, J.P. , Cantarel, A.A.M. , Simon, L. & Haichar, F.E.Z. (2018) Root exudation rate as functional trait involved in plant nutrient‐use strategy classification. Ecology and Evolution, 8, 8573–8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyssels, G. , Poesen, J. , Bochet, E. & Li, Y. (2005) Impact of plant roots on the resistance of soils to erosion by water: a review. Progress in Physical Geography: Earth and Environment, 29, 189–217. [Google Scholar]
- Haling, R.E. , Simpson, R.J. , McKay, A.C. , Hartley, D. , Lambers, H. & Richardson, A.E. (2011) Direct measurement of roots in soil for single and mixed species using a quantitative DNA‐based method. Plant and Soil, 348, 123–137. [Google Scholar]
- Hallama, M. , Pekrun, C. , Lambers, H. & Kandeler, E. (2019). Hidden miners–the roles of cover crops and soil microorganisms in phosphorus cycling through agroecosystems. Plant and Soil, 435, 7–45. [Google Scholar]
- Han, E. , Kautz, T. , Perkons, U. , Lüsebrink, M. , Pude, R. & Köpke, U. (2015) Quantification of soil biopore density after perennial fodder cropping. Plant and Soil, 394, 73–85. [Google Scholar]
- Han, E. , Li, F. , Perkons, U. , Küpper, P.M. , Bauke, S.L. , Athmann, M. et al. (2021) Can precrops uplift subsoil nutrients to topsoil? Plant and Soil, 463, 329–345. [Google Scholar]
- Hansen, V. , Eriksen, J. , Jensen, L.S. , Thorup‐Kristensen, K. & Magid, J. (2021) Towards integrated cover crop management: N, P and S release from aboveground and belowground residues. Agriculture, Ecosystems & Environment, 313, 107392. [Google Scholar]
- Hao, L. , Pu, X. & Song, J. (2021) Introduction of mutations in plants with prime editing. Methods, 194, 83–93. [DOI] [PubMed] [Google Scholar]
- Hao, L. , Zhang, Z. , Hao, B. , Diao, F. , Zhang, J. , Bao, Z. et al. (2021) Arbuscular mycorrhizal fungi alter microbiome structure of rhizosphere soil to enhance maize tolerance to La. Ecotoxicology and Environmental Safety, 212, 111996. [DOI] [PubMed] [Google Scholar]
- Haramoto, E.R. & Gallandt, E.R. (2004) Brassica cover cropping for weed management: a review. Renewable Agriculture and Food Systems, 19, 187–198. [Google Scholar]
- Haudry, A. , Cenci, A. , Ravel, C. , Bataillon, T. , Brunel, D. , Poncet, C. et al. (2007) Grinding up wheat: a massive loss of nucleotide diversity since domestication. Molecular Biology and Evolution, 24, 1506–1517. [DOI] [PubMed] [Google Scholar]
- Hector, A. , Schmid, B. , Beierkuhnlein, C. , Caldeira, M.C. , Diemer, M. , Dimitrakopoulos, P.G. et al. (1999) Plant diversity and productivity experiments in european grasslands. Science, 286, 1123–1127. [DOI] [PubMed] [Google Scholar]
- Helliwell, J.R. , Sturrock, C.J. , Mairhofer, S. , Craigon, J. , Ashton, R.W. , Miller, A.J. et al. (2017) The emergent rhizosphere: imaging the development of the porous architecture at the root‐soil interface. Scientific Reports, 7, 14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, D.R. , Riga, E. , Ramirez, R.A. , Wilson, J. & Snyder, W.E. (2009) Mustard biofumigation disrupts biological control by Steinernema spp. nematodes in the soil. Biological Control, 48, 316–322. [Google Scholar]
- Henneron, L. , Cros, C. , Picon‐Cochard, C. , Rahimian, V. & Fontaine, S. (2020) Plant economic strategies of grassland species control soil carbon dynamics through rhizodeposition. The Journal of Ecology, 108, 528–545. [Google Scholar]
- Henneron, L. , Kardol, P. , Wardle, D.A. , Camille, C. , Fontaine, S. (2020) Rhizosphere control of soil nitrogen cycling: a key component of plant economic strategies. The New Phytologist, 228, 1269–1282. [DOI] [PubMed] [Google Scholar]
- Hensel, G. , Kastner, C. , Oleszczuk, S. , Riechen, J. & Kumlehn, J. (2009) Agrobacterium‐mediated gene transfer to cereal crop plants: current protocols for barley, wheat, triticale, and maize. International Journal of Plant Genomics, 2009, 835608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herridge, D. & Rose, I. (2000) Breeding for enhanced nitrogen Æxation in crop legumes. Field Crops Research, 65, 229–248. [Google Scholar]
- Heuermann, D. , Gentsch, N. , Boy, J. , Schweneker, D. , Feuerstein, U. , Groß, J. et al. (2019) Interspecific competition among catch crops modifies vertical root biomass distribution and nitrate scavenging in soils. Scientific Reports, 9, 11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey, L.T. , N Hafeez, A. , Robinson, H. , Jackson, S.A. , Leal‐Bertioli, S.C.M. , Tester, M. et al. (2019) Breeding crops to feed 10 billion. Nature Biotechnology, 37, 744–754. [DOI] [PubMed] [Google Scholar]
- Hinsinger, P. , Betencourt, E. , Bernard, L. , Brauman, A. , Plassard, C. , Shen, J. et al. (2011) P for two, sharing a scarce resource: soil phosphorus acquisition in the rhizosphere of intercropped species. Plant Physiology, 156, 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, M.D. , Rosas, J.C. , Brown, K.M. & Lynch, J.P. (2005) Root architectural tradeoffs for water and phosphorus acquisition. Functional Plant Biology, 32, 737–748. [DOI] [PubMed] [Google Scholar]
- Holme, I.B. , Gregersen, P.L. & Brinch‐Pedersen, H. (2019) Induced genetic variation in crop plants by random or targeted mutagenesis: convergence and differences. Frontiers in Plant Science, 10, 1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, L. , Robert, C.A.M. , Cadot, S. , Zhang, X. , Ye, M. , Li, B. et al. (2018) Root exudate metabolites drive plant‐soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nature Communications, 9, 2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, N. , Athmann, M. & Han, E. (2020) Biopore‐induced deep root traits of two winter crops. Collection FAO: Agriculture, 10, 634. [Google Scholar]
- Hunter, M.C. , Smith, R.G. , Schipanski, M.E. , Atwood, L.W. & Mortensen, D.A. (2017) Agriculture in 2050: recalibrating targets for sustainable intensification. Bioscience, 67, 386–391. [Google Scholar]
- Hyten, D.L. , Song, Q. , Zhu, Y. , Choi, I.‐Y. , Nelson, R.L. , Costa, J.M. et al. (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proceedings of the National Academy of Sciences of the United States of America, 103, 16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- in 't Zandt, D. , Hoekstra, N.J. , Wagemaker, C.A.M. , Caluwe, H. , Smit‐Tiekstra, A.E. , Visser, E.J.W. et al. (2020) Local soil legacy effects in a multispecies grassland community are underlain by root foraging and soil nutrient availability. The Journal of Ecology, 108, 2243–2255. [Google Scholar]
- Jankowicz‐Cieslak, J. , Mba, C. & Till, B.J. (2017) Mutagenesis for crop breeding and functional genomics. Biotechnologies for Plant Mutation Breeding, 30, 3–18. [Google Scholar]
- Jarvis, B.A. , Romsdahl, T.B. , McGinn, M.G. , Nazarenus, T.J. , Cahoon, E.B. , Chapman, K.D. et al. (2021) CRISPR/Cas9‐Induced fad2 and rod1 mutations stacked with fae1 confer high oleic acid seed oil in pennycress (Thlaspi arvense L.). Frontiers in Plant Science, 12, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. & Charpentier, E. (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C. , de Vega, J. , Lloyd, D. , Hegarty, M. , Ayling, S. , Powell, W. et al. (2020) Population structure and genetic diversity in red clover (Trifolium pratense L.) germplasm. Scientific Reports, 10, 8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar, T.C. & Singer, J.W. (2011) The use of cover crops to manage soil, In: Hatfield, J.L., Sauer, T.J. (Eds.) Soil Management: Building a Stable Base for Agriculture. Madison, WI: Soil Science Society of America, pp. 321–337. [Google Scholar]
- Kautz, T. , Amelung, W. , Ewert, F. , Gaiser, T. , Horn, R. , Jahn, R. et al. (2013) Nutrient acquisition from arable subsoils in temperate climates: a review. Soil Biology and Biochemistry, 57, 1003–1022. [Google Scholar]
- Kilburn, M.R. , Jones, D.L. , Clode, P.L. , Cliff, J.B. , Stockdale, E.A. , Herrmann, A.M. et al. (2010) Application of nanoscale secondary ion mass spectrometry to plant cell research. Plant Signaling & Behavior, 5, 760–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, M. , Arai, Y. , Iwase, N. , Shirotori, K. , Shioiri, H. & Nozue, M. (2000) Development of a simple and efficient method for transformation of buckwheat plants (Fagopyrum esculentum) using Agrobacterium tumefaciens. Bioscience, Biotechnology, and Biochemistry, 64, 845–847. [DOI] [PubMed] [Google Scholar]
- Kristensen, H.L. & Thorup‐Kristensen, K. (2004) Root growth and nitrate uptake of three different catch crops in deep soil layers. Soil Science Society of America Journal, 68, 529–537. [Google Scholar]
- Kurowska, M. , Daszkowska‐Golec, A. , Gruszka, D. , Marzec, M. , Szurman, M. , Szarejko, I. et al. (2011) TILLING‐a shortcut in functional genomics. Journal of Applied Genetics, 52, 371–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, H.M. , Murray, S.C. , Montesinos‑López, O.A. , Montesinos‑López, A. , Crossa, J. , Rooney, et al. (2020) Phenomic selection and prediction of maize grain yield from near‐infrared reflectance spectroscopy of kernels. The Plant Phenome Journal, 3, e20002. [Google Scholar]
- Langdale, G.W. , Blevins, R.L. , Karlen, D.L. , McCool, D.K. , Nearing, M.A. , Skidmore, E.L. et al. (1991) Cover crop effects on soil erosion by wind and water, In: Hargrove, W.L. (Eds.) Cover Crops for Clean Water. Ankeny, Iowa: Soil and Water Conservation Society, pp. 15–22. https://www.ars.usda.gov/ARSUserFiles/30200525/91–%20Cover%20Crop%20Effects%20on%20Soil%20Erosion.pdf [Google Scholar]
- Lehmann, J. , Bossio, D.A. , Kögel‐Knabner, I. & Rillig, M.C. (2020) The concept and future prospects of soil health. Nature Reviews Earth and Environment, 1, 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Li, J. , Chen, J. , Yan, L. & Xia, L. (2020) Precise modifications of both exogenous and endogenous genes in rice by prime editing. Molecular Plant, 13, 671–674. [DOI] [PubMed] [Google Scholar]
- Li, J. , Sun, Y. , Du, J. , Zhao, Y. & Xia, L. (2017) Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Molecular Plant, 10, 526–529. [DOI] [PubMed] [Google Scholar]
- Lin, Q. , Jin, S. , Zong, Y. , Yu, H. , Zhu, Z. , Liu, G. et al. (2021) High‐efficiency prime editing with optimized, paired pegRNAs in plants. Nature Biotechnology, 39, 923–927. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Dong, X. , Xue, Q. , Leskovar, D.I. , Jifon, J. , Butnor, J.R. et al. (2018) Ground penetrating radar (GPR) detects fine roots of agricultural crops in the field. Plant and Soil, 423, 517–531. [Google Scholar]
- Liu, Y. , Cui, Z. , Huang, Z. , López‐Vicente, M. & Wu, G.‐L. (2019) Influence of soil moisture and plant roots on the soil infiltration capacity at different stages in arid grasslands of China. Catena, 182, 104147. [Google Scholar]
- Loreau, M. & Hector, A. (2001) Partitioning selection and complementarity in biodiversity experiments. Nature, 412, 72–76. [DOI] [PubMed] [Google Scholar]
- Louvieaux, J. , Spanoghe, M. & Hermans, C. (2020) Root morphological traits of seedlings are predictors of seed yield and quality in winter oilseed rape hybrid cultivars. Frontiers in Plant Science, 11, 568009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder, L.G. , Zhang, D. , Baltes, N.J. , Paul, J.W. 3rd , Tang, X. , Zheng, X. & Qi, Y. (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiology, 169, 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. et al. (2015) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot Plants. Molecular Plant, 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Macias‐Benitez, S. , Garcia‐Martinez, A.M. , Caballero Jimenez, P. , Gonzalez, J.M. , Tejada Moral, M. , Parrado et al. (2020) Rhizospheric organic acids as biostimulants: monitoring feedbacks on soil microorganisms and biochemical properties. Frontiers in plant science, 11, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdoff, F. & van Es, H. (2009) Building soils for better crops: sustainable soil management. Brentwood, MD: SARE Outreach Publications c/o International Fulfillment Corporation.
- Malhi, S.S. , Grant, C.A. , Johnston, A.M. & Gill, K.S. (2001) Nitrogen fertilization management for no‐till cereal production in the Canadian Great Plains: a review. Soil and Tillage Research, 60, 101–122. [Google Scholar]
- Marks, M.D. , Chopra, R. & Sedbrook, J.C. (2021) Technologies enabling rapid crop improvements for sustainable agriculture: example pennycress (Thlaspi arvense L.). Emerging Topics in Life Sciences, 5, 325–335. [DOI] [PubMed] [Google Scholar]
- Mergoum, M. , Singh, P.K. , Pena, R.J. , Lozano‐del Rio, A.J. , Cooper, K.V. , Salmon, D.F. et al. (2009) Triticale: a “New” crop with old challenges. In: Carena, M.J. (Eds.) Cereals. New York, NY: Springer. pp. 267–287. [Google Scholar]
- McDaniel, M.D. , Tiemann, L.K. & Grandy, A.S. (2014) Does agricultural crop diversity enhance soil microbial biomass and organic matter dynamics? A meta‐analysis. Ecological Applications, 24, 560–570. [DOI] [PubMed] [Google Scholar]
- McGinn, M. , Phippen, W.B. , Chopra, R. , Bansal, S. , Jarvis, B.A. , Phippen, M.E. et al. (2019) Molecular tools enabling pennycress (Thlaspi arvense) as a model plant and oilseed cash cover crop. Plant Biotechnology Journal, 17, 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle, T.P. , Miller, M.H. , Evans, D.G. , Fairchild, G.L. & Swan, J.A. (1990) A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytologist, 115, 495–501. [DOI] [PubMed] [Google Scholar]
- McPherson, M.R. , Wang, P. , Marsh, E.L. , Mitchell, R.B. & Schachtman, D.P. (2018) Isolation and analysis of microbial communities in soil, rhizosphere, and roots in perennial grass experiments. Journal of Visualized Experiments, 137, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinen, C. & Rauber, R. (2015) Root discrimination of closely related crop and weed species using FT MIR‐ATR spectroscopy. Frontiers in Plant Science, 6, 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, S.T. , Ptacnik, R. , Hillebrand, H. , Bessler, H. , Buchmann, N. , Ebeling, A. et al. (2018) Biodiversity–multifunctionality relationships depend on identity and number of measured functions. Nature Ecology & Evolution, 2, 44–49. [DOI] [PubMed] [Google Scholar]
- Mishra, R. , Joshi, R.K. & Zhao, K. (2020) Base editing in crops: current advances, limitations and future implications. Plant Biotechnology Journal, 18, 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommer, L. , Van Ruijven, J. , De Caluwe, H. , Smit‐Tiekstra, A.E. , Wagemaker, C.A.M. , Joop Ouborg, N. et al. (2010) Unveiling below‐ground species abundance in a biodiversity experiment: a test of vertical niche differentiation among grassland species. The Journal of Ecology, 98, 1117–1127. [Google Scholar]
- Mommer, L. , Wagemaker, C.A.M. , De Kroon, H. & Ouborg, N.J. (2008) Unravelling below‐ground plant distributions: a real‐time polymerase chain reaction method for quantifying species proportions in mixed root samples. Molecular Ecology Resources, 8, 947–953. [DOI] [PubMed] [Google Scholar]
- Morris, E.C. , Griffiths, M. , Golebiowska, A. , Mairhofer, S. , Burr‐Hersey, J. & Goh, T. et al. (2017) Shaping 3D root system architecture. Current Biology, 27, R919–R930. [DOI] [PubMed] [Google Scholar]
- Nadolska‐Orczyk, A. , Przetakiewicz, A. , Kopera, K. , Binka, A. & Orczyk, W. (2005) Efficient method of agrobacterium‐mediated transformation for Triticale (x Triticosecale Wittmack). Journal of Plant Growth Regulation, 24, 2–10. [Google Scholar]
- Nagel, K.A. , Putz, A. , Gilmer, F. , Heinz, K. , Fischbach, A. & Pfeifer, J. et al. (2012) GROWSCREEN‐Rhizo is a novel phenotyping robot enabling simultaneous measurements of root and shoot growth for plants grown in soil‐filled rhizotrons. Functional Plant Biology, 39, 891–904. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. & Searle, I.R. (2022). An efficient root transformation system for recalcitrant Vicia sativa. Frontiers in Plant Science, 12, 781014. 10.3389/fpls.2021.781014 [DOI] [PMC free article] [PubMed]
- Nunes, M.R. , van Es, H.M. , Schindelbeck, R. , Ristow, A.J. & Ryan, M. (2018) No‐till and cropping system diversification improve soil health and crop yield. Geoderma, 328, 30–43. [Google Scholar]
- Oburger, E. & Jones, D.L. (2018) Sampling root exudates—mission impossible? Rhizosphere, 6, 116–133. [Google Scholar]
- Oettler, G. , Wehmann, F. & Utz, H.F. (1991) Influence of wheat and rye parents on agronomic characters in primary hexaploid and octoploid triticale. Theoretical and Applied Genetics, 81, 401–405. [DOI] [PubMed] [Google Scholar]
- Osipitan, O.A. , Dille, J.A. , Assefa, Y. , Radicetti, E. , Ayeni, A. & Knezevic, S.Z. (2019) Impact of cover crop management on level of weed suppression: a meta‐analysis. Crop Science, 59, 833–842. [Google Scholar]
- Pace, G.M. & McClure, P.R. (1986) Comparison of nitrate uptake kinetic parameters across maize inbred lines. Journal of Plant Nutrition, 9, 1095–1111. [Google Scholar]
- Paez‐Garcia, A. , Motes, C.M. , Scheible, W.‐R. , Chen, R. , Blancaflor, E.B. & Monteros, M.J. (2015) Root traits and phenotyping strategies for plant improvement. Plants, 4, 334–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, B.K. , Huang, G. , Bhosale, R. , Hartman, S. , Sturrock, C.J. , Jose, L. et al. (2021) Plant roots sense soil compaction through restricted ethylene diffusion. Science, 371, 276–280. [DOI] [PubMed] [Google Scholar]
- Pang, J. , Bansal, R. , Zhao, H. , Bohuon, E. , Lambers, H. , Ryan, M.H. , et al. (2018) The carboxylate‐releasing phosphorus‐mobilizing strategy can be proxied by foliar manganese concentration in a large set of chickpea germplasm under low phosphorus supply. New Phytologist, 219, 518–529. [DOI] [PubMed] [Google Scholar]
- Pang, R. , Xu, X. , Tian, Y. , Cui, X. , Ouyang, H. & Kuzyakov, Y. (2021) In‐situ 13CO2 labeling to trace carbon fluxes in plant‐soil‐microorganism systems: review and methodological guideline. Rhizosphere, 20, 100441. [Google Scholar]
- Pflugfelder, D. , Metzner, R. , van Dusschoten, D. , Reichel, R. , Jahnke, S. & Koller, R. (2017) Non‐invasive imaging of plant roots in different soils using magnetic resonance imaging (MRI). Plant Methods, 13, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda, A. , Kaplan, I. , Hannula, S.E. , Ghanem, W. & Bezemer, T.M. (2020) Conditioning the soil microbiome through plant‐soil feedbacks suppresses an aboveground insect pest. The New Phytologist, 226, 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires, L.F. , Roque, W.L. , Rosa, J.A. & Mooney, S.J. (2019) 3D analysis of the soil porous architecture under long term contrasting management systems by X‐ray computed tomography. Soil and Tillage Research, 191, 197–206. [Google Scholar]
- Postma, J.A. & Lynch, J.P. (2012) Complementarity in root architecture for nutrient uptake in ancient maize/bean and maize/bean/squash polycultures. Annals of Botany, 110, 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuters (2021) Corteva, Indigo Ag team up on carbon credit program for U.S. farmers. Available at: https://www.reuters.com/business/sustainable-business/corteva-indigo-ag-team-up-carbon-credit-program-us-farmers-2021-08-26/ [Accessed 11th April 2021].
- Rewald, B. , Meinen, C. , Trockenbrodt, M. , Ephrath, J.E. & Rachmilevitch, S. (2012) Root taxa identification in plant mixtures—current techniques and future challenges. Plant and Soil, 359, 165–182. [Google Scholar]
- Reynolds, M. , Atkin, O.K. , Bennett, M. , Cooper, M. , Dodd, I.C. , Foulkes, M.J. et al. (2021) Addressing research bottlenecks to crop productivity. Trends in Plant Science, 26, 607–630. [DOI] [PubMed] [Google Scholar]
- Reynolds, M. , Foulkes, M.J. , Slafer, G.A. , Berry, P. , Parry, M.A.J. , Snape, J.W. et al. (2009) Raising yield potential in wheat. Journal of Experimental Botany, 60, 1899–1918. [DOI] [PubMed] [Google Scholar]
- Riday, H. & Krohn, A.L. (2010) Genetic map‐based location of the red clover (Trifolium pratense L.) gametophytic self‐incompatibility locus. Theoretical and Applied Genetics, 121, 761–767. [DOI] [PubMed] [Google Scholar]
- Rincent, R. , Charpentier, J.‐P. , Faivre‐Rampant, P. , Paux, E. , Le Gouis, J. , Bastien, C. et al. (2018) Phenomic selection is a low‐cost and high‐throughput method based on indirect predictions: proof of concept on wheat and poplar. G3, 8, 3961–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Leal, D. , Lemmon, Z.H. , Man, J. , Bartlett, M.E. & Lippman, Z.B. (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell, 171, 470–480. [DOI] [PubMed] [Google Scholar]
- Roy, S. , Liu, W. , Nandety, R.S. , Crook, A. , Mysore, K.S. , Pislariu, C.I. et al. (2020) Celebrating 20 years of genetic discoveries in Legume nodulation and symbiotic nitrogen fixation. The Plant Cell, 32, 15–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, N.Z. , dos Dieckow, J. , Bayer, C. , Molin, R. , Favaretto, N. , Pauletti, V. et al. (2011) Forages, cover crops and related shoot and root additions in no‐till rotations to C sequestration in a subtropical ferralsol. Soil and Tillage Research, 111, 208–218. [Google Scholar]
- Sasse, J. , Martinoia, E. & Northen, T. (2018) Feed your friends: do plant exudates shape the root microbiome? Trends in Plant Science, 23, 25–41. [DOI] [PubMed] [Google Scholar]
- Seethepalli, A. , Dhakal, K. , Griffiths, M. , Guo, H. , Freschet, G.T. & York, L.M. (2021) RhizoVision explorer: open‐source software for root image analysis and measurement standardization. AoB Plants, 13, plab056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seethepalli, A. , Guo, H. , Liu, X. , Griffiths, M. , Almtarfi, H. , Li, Z. , et al. (2020) RhizoVision crown: an integrated hardware and software platform for root crown phenotyping. Plant Phenomics, 2020, 3074916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert, C.A. , Roberts, M.J. & Lobell, D.B. (2017) Continuous corn and soybean yield penalties across hundreds of thousands of fields. Agronomy Journal, 109, 541–548. [Google Scholar]
- Shane, M.W. & Lambers, H. (2005) Cluster roots: a curiosity in context. Plant and Soil, 274, 101–125. [Google Scholar]
- Shen, J. , Rengel, Z. , Tang, C. & Zhang, F. (2003) Role of phosphorus nutrition in development of cluster roots and release of carboxylates in soil‐grown Lupinus albus . Plant and Soil, 248, 199–206. [Google Scholar]
- Smith, A.G. , Han, E. , Petersen, J. , Olsen, N.A.F. , Giese, C. & Athmann, M. et al. (2022). RootPainter: Interactive‐machine‐learning enables rapid and accurate contouring for radiotherapy. Medical Physics, 49, 461–473. 10.1002/mp.15353 [DOI] [PubMed] [Google Scholar]
- Smýkal, P. , Aubert, G. , Burstin, J. , Coyne, C.J. , Ellis, N.T.H. , Flavell, A.J. et al. (2012) Pea (Pisum sativum L.) in the Genomic EraPea (Pisum sativum L.) in the Genomic Era. Agronomy, 2, 74–115. [Google Scholar]
- Soltaninejad, M. , Sturrock, C.J. , Griffiths, M. , Pridmore, T.P. & Pound, M.P. (2020) Three dimensional root CT segmentation using multi‐resolution encoder‐decoder networks. IEEE Transactions on Image Processing, 29, 6667–6679. [DOI] [PubMed] [Google Scholar]
- Sun, B. , Gao, Y. & Lynch, J.P. (2018) Large crown root number improves topsoil foraging and phosphorus acquisition. Plant Physiology, 177, 90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Ataka, M. , Han, M. , Han, Y. , Gan, D. , Xu, T. et al. (2021) Root exudation as a major competitive fine‐root functional trait of 18 coexisting species in a subtropical forest. The New Phytologist, 229, 259–271. [DOI] [PubMed] [Google Scholar]
- Tang, X. , Sretenovic, S. , Ren, Q. , Jia, X. , Li, M. , Fan, T. , et al. (2020) Plant prime editors enable precise gene editing in rice cells. Molecular Plant, 13, 667–670. [DOI] [PubMed] [Google Scholar]
- Tani, E. , Abraham, E. , Chachalis, D. & Travlos, I. (2017) Molecular, genetic and agronomic approaches to utilizing pulses as cover crops and green manure into cropping systems. International Journal of Molecular Sciences, 18, 1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wall Street Journal (2021) U.S. farmers look for government help to support Biden's climate plans. Available at: https://www.wsj.com/articles/u-s-farmers-look-for-government-help-to-support-bidens-climate-plans-11619861401 [Accessed 20th May 2021].
- Thomas, C.L. , Graham, N.S. , Hayden, R. , Meacham, M.C. , Neugebauer, K. , Nightingale, M. et al. (2016) High‐throughput phenotyping (HTP) identifies seedling root traits linked to variation in seed yield and nutrient capture in field‐grown oilseed rape (Brassica napus L.). Annals of Botany, 118, 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorup‐Kristensen, K. , Dresbøll, D.B. & Kristensen, H.L. (2012) Crop yield, root growth, and nutrient dynamics in a conventional and three organic cropping systems with different levels of external inputs and N re‐cycling through fertility building crops. European Journal of Agronomy, 37, 66–82. [Google Scholar]
- Thorup‐Kristensen, K. , Magid, J. & Jensen, L.S. (2003) Catch crops and green manures as biological tools in nitrogen management in temperate zones. In: Sparks, D. (Eds.) Advances in Agronomy. Academic Press. pp. 227–302 [Google Scholar]
- Tiemann, L.K. , Grandy, A.S. , Atkinson, E.E. , Marin‐Spiotta, E. & McDaniel, M.D. (2015) Crop rotational diversity enhances belowground communities and functions in an agroecosystem. Ecology Letters, 18, 761–771. [DOI] [PubMed] [Google Scholar]
- Tittonell, P. (2014) Ecological intensification of agriculture—sustainable by nature. Current Opinion in Environmental Sustainability, 8, 53–61. [Google Scholar]
- Topp, C.N. , Bray, A.L. , Ellis, N.A. & Liu, Z. (2016) How can we harness quantitative genetic variation in crop root systems for agricultural improvement? Journal of Integrative Plant Biology, 58, 213–225. [DOI] [PubMed] [Google Scholar]
- Torbert, H.A. , Reeves, D.W. & Mulvaney, R.L. (1996) Winter legume cover crop benefits to corn: rotation vs. fixed‐nitrogen effects. Agronomy Journal, 88, 527–535. [Google Scholar]
- Trachsel, S. , Kaeppler, S.M. , Brown, K.M. & Lynch, J.P. (2011) Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant and Soil, 341, 75–87. [Google Scholar]
- Trachsel, S. , Kaeppler, S.M. , Brown, K.M. & Lynch, J.P. (2013) Maize root growth angles become steeper under low N conditions. Field Crops Research, 140, 18–31. [Google Scholar]
- Tracy, S.R. , Black, C.R. , Roberts, J.A. , Dodd, I.C. & Mooney, S.J. (2015) Using X‐ray computed tomography to explore the role of abscisic acid in moderating the impact of soil compaction on root system architecture. Environmental and Experimental Botany, 110, 11–18. [Google Scholar]
- Uga, Y. , Sugimoto, K. , Ogawa, S. , Rane, J. , Ishitani, M. , Hara, N. et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nature Genetics, 45, 1097–1102. [DOI] [PubMed] [Google Scholar]
- van der Putten, W.H. , Bardgett, R.D. , Bever, J.D. , Bezemer, T.M. , Casper, B.B. , Fukami, T. et al. (2013) Plant‐soil feedbacks: the past, the present and future challenges. The Journal of Ecology, 101, 265–276. [Google Scholar]
- van Noordwijk, M. , Brouwer, G. , Meijboom, F. , do Rosário, G. , Oliveira, M. & Bengough, A.G. (2001) Trench profile techniques and core break methods. Root Methods, 8, 211–233. [Google Scholar]
- Vats, S. , Kumawat, S. , Kumar, V. , Patil, G.B. , Joshi, T. , Sonah, H. et al. (2019) Genome editing in plants: exploration of technological advancements and challenges. Cells, 8, 1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives‐Peris, V. , de Ollas, C. , Gómez‐Cadenas, A. & Pérez‐Clemente, R.M. (2020) Root exudates: from plant to rhizosphere and beyond. Plant Cell Reports, 39, 3–17. [DOI] [PubMed] [Google Scholar]
- de Vries, F.T. , Williams, A. , Stringer, F. , Willcocks, R. , McEwing, R. , Langridge, H. et al. (2019) Changes in root‐exudate‐induced respiration reveal a novel mechanism through which drought affects ecosystem carbon cycling. The New Phytologist, 224, 132–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukicevich, E. , Lowery, T. , Bowen, P. , Úrbez‐Torres, J.R. & Hart, M. (2016) Cover crops to increase soil microbial diversity and mitigate decline in perennial agriculture. A review. Agronomy for Sustainable Development, 36, 48. [Google Scholar]
- Wagemaker, C.A.M. , Mommer, L. , Visser, E.J.W. , Weigelt, A. , van Gurp, T.P. , Postuma, M. et al. (2021) msGBS: a new high‐throughput approach to quantify the relative species abundance in root samples of multispecies plant communities. Molecular Ecology Resources, 21, 1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, C. (2005) A simple blue staining technique for arbuscular mycorrhizal and other root‐inhabiting fungi. Inoculum, 56, 68–69. [Google Scholar]
- Wang, N.‐Q. , Kong, C.‐H. , Wang, P. & Meiners, S.J. (2021) Root exudate signals in plant‐plant interactions. Plant, Cell & Environment, 44, 1044–1058. [DOI] [PubMed] [Google Scholar]
- Wang, N.‐Q. , Mathews, A.J. , Li, K. , Wen, J. , Komarov, S. , O'Sullivan, J.A. et al. (2014) A dedicated high‐resolution PET imager for plant sciences. Physics in Medicine and Biology, 59, 5613–5629. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Chen, L. & Ma, J. (2019) Genomic introgression through interspecific hybridization counteracts genetic bottleneck during soybean domestication. Genome Biology, 20, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson, A. , Bischof, L. , Zwart, A. & Watt, M. (2016) A portable fluorescence spectroscopy imaging system for automated root phenotyping in soil cores in the field. Journal of Experimental Botany, 67, 1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidlich, E.W.A. , Temperton, V.M. & Faget, M. (2018) Neighbourhood stories: role of neighbour identity, spatial location and order of arrival in legume and non‐legume initial interactions. Plant and Soil, 424, 171–182. [Google Scholar]
- Weigand, M. & Kemna, A. (2017) Multi‐frequency electrical impedance tomography as a non‐invasive tool to characterize and monitor crop root systems. Biogeosciences, 14, 921–939. [Google Scholar]
- Weisser, W.W. , Roscher, C. , Meyer, S.T. , Ebeling, A. , Luo, G. , Allan, E. et al. (2017) Biodiversity effects on ecosystem functioning in a 15‐year grassland experiment: patterns, mechanisms, and open questions. Basic and Applied Ecology, 23, 1–73. [Google Scholar]
- Wen, Z. , Li, H. , Shen, Q. , Tang, X. , Xiong, C. , Li, H. , Pang, J. , Ryan, M.H. , Lambers, H. & Shen, J . (2019). Tradeoffs among root morphology, exudation and mycorrhizal symbioses for phosphorus‐acquisition strategies of 16 crop species. New Phytologist, 223(2), 882–895. 10.1111/nph.15833 [DOI] [PubMed] [Google Scholar]
- White, C.M. , DuPont, S.T. , Hautau, M. , Hartman, D. , Finney, D.M. , Bradley, B. , et al. (2017) Managing the trade off between nitrogen supply and retention with cover crop mixtures. Agriculture, Ecosystems and Environment, 237, 121–133. [Google Scholar]
- Williams, A. & de Vries, F.T. (2020) Plant root exudation under drought: implications for ecosystem functioning. The New Phytologist, 225, 1899–1905. [DOI] [PubMed] [Google Scholar]
- Williams, A. , Langridge, H. , Straathof, A.L. , Fox, G. , Muhammadali, H. , Hollywood, K.A. , et al. (2021a) Comparing root exudate collection techniques: an improved hybrid method. Soil Biology and Biochemistry, 161, 108391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, A. , Langridge, H. , Straathof, A.L. , Muhamadali, H. , Hollywood, K.A. & Goodacre, R. et al. (2021b) Root functional traits explain root exudation rate and composition across a range of grassland species. The Journal of Ecology, 13630, 1365–2745. 10.1111/1365-2745.13630 [DOI] [Google Scholar]
- Wittwer, R.A. , Dorn, B. , Jossi, W. & van der Heijden, M.G.A. (2017) Cover crops support ecological intensification of arable cropping systems. Scientific Reports, 7, 41911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer, R.A. & van der Heijden, M.G.A. (2020) Cover crops as a tool to reduce reliance on intensive tillage and nitrogen fertilization in conventional arable cropping systems. Field Crops Research, 249, 107736. [Google Scholar]
- Wood, S.A. & Bowman, M. (2021) Large‐scale farmer‐led experiment demonstrates positive impact of cover crops on multiple soil health indicators. Nature Food, 2, 97–103. [DOI] [PubMed] [Google Scholar]
- Xing, H.‐L. , Dong, L. , Wang, Z.‐P. , Zhang, H.‐Y. , Han, C.‐Y. , Liu, B. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biology, 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.J. , Samayoa, L.F. , Bradbury, P.J. , Olukolu, B.A. , Xue, W. , York, A.M. et al. (2019) The genetic architecture of teosinte catalyzed and constrained maize domestication. Proceedings of the National Academy of Sciences of the United States of America, 116, 5643–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, L.M. , Carminati, A. , Mooney, S.J. , Ritz, K. & Bennett, M.J. (2016) The holistic rhizosphere: integrating zones, processes, and semantics in the soil influenced by roots. Journal of Experimental Botany, 67, 3629–3643. [DOI] [PubMed] [Google Scholar]
- Zhalnina, K. , Louie, K.B. , Hao, Z. , Mansoori, N. , da Rocha, U.N. , Shi, S. et al. (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nature Microbiology, 3, 470–480. [DOI] [PubMed] [Google Scholar]
- Zhan, A. & Lynch, J.P. (2015) Reduced frequency of lateral root branching improves N capture from low‐N soils in maize. Journal of Experimental Botany, 66, 2055–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Whalley, W.R. , Hawkesford, M.J. , Ashton, R.W. , Atkinson, B. , Atkinson, J.A. et al. (2021) The interaction between wheat roots and soil pores in structured field soil. Journal of Experimental Botany, 72, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J. , Ingram, P.A. , Benfey, P.N. & Elich, T. (2011) From lab to field, new approaches to phenotyping root system architecture. Current Opinion in Plant Biology, 14, 310–317. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Kaeppler, S.M. & Lynch, J.P. (2005) Topsoil foraging and phosphorus acquisition efficiency in maize (Zea mays). Functional Plant Biology, 32, 749–762. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Chen, Y. , Ali, A. , Dong, L. , Wang, X. , Archontoulis, S.V. et al. (2021) Continuous in situ soil nitrate sensors: the importance of high‐resolution measurements across time and a comparison with salt extraction‐based methods. Soil Science Society of America Journal, 2021, 1–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Images, data, and statistical analysis scripts available at 10.5281/zenodo.5039308


