Figure 6.
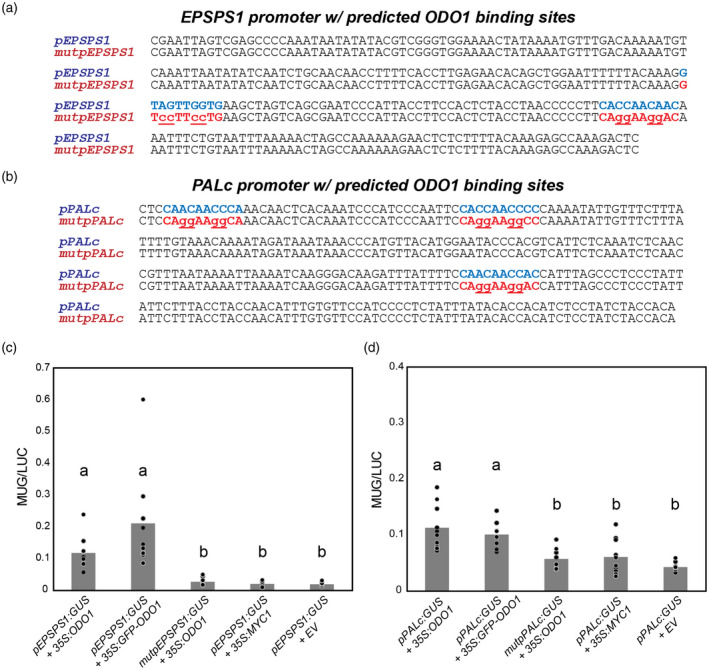
Activation of EPSPS1 and PALc promoters by ODO1 in N. benthamiana. Promoter regions immediately preceding the predicted transcription start site and containing predicted ODO1 binding sites from MEME (see also Figure S7) in blue for (a) EPSPS1 and (b) PALc. Binding sites in pEPSPS1 and pPALC were mutated to form mutpEPSPS1 and mutpPALc constructs as shown (mutated bases lowercase, underlined). N. benthamiana leaves were infiltrated with A. tumefaciens harboring the effector constructs 35S:ODO1, 35S:MYC1, 35S:GFP‐ODO1, or empty vector (pBINPLUS, EV) and the reporter constructs (c) pEPSPS1:GUS or mutpEPSPS1:GUS or (d) pPALc:GUS or mutpPALc:GUS. GUS activity was normalized with 35S:LUC activity. For both promoter constructs, in the presence of ODO1, mutation of predicted ODO1 binding sites led to reduction of promoter GUS activity to background levels in the range observed for MYC1 or empty vector. Tagged GFP‐ODO1 as used in ChIP‐Seq showed similar activity to untagged protein. Bars that do not share a letter have a significant difference determined by Kruskal‐Wallis test and pairwise comparisons adjusted by Bonferroni correction for multiple tests (P < 0.05). Individual data points are depicted with dots. Abbreviations: EPSPS, 5‐enolpyruvylshikimate 3‐phosphate; GUS, β‐glucuronidase; LUC, luciferase; ODO1, ODORANT1; PAL, phenylalanine ammonia lyase.
