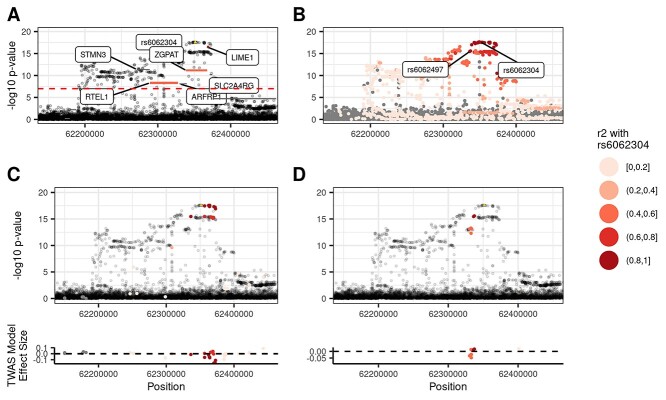
Figure 6.
TWAS and coloc variant-to-gene assignments agree at LIME1 locus. The LIME1-lymphocyte percentage associated locus illustrates one example where the TWAS- and coloc-based variant-to-gene assignments agree. (A) Genes in the FINEMAP credible set are colored by their correlation with rs6062304. The predicted gene expression with LIME1 is highly correlated with rs6062304, whereas neither ZGPAT nor RTEL1 are. (B) An eQTL for LIME1, rs6062497, is in high LD with rs6062304, and in turn coloc assigns LIME1 as an eGene. (C, D) Model variants for LIME1 and ZGPAT are colored by their LD with rs6062304, respectively. The variants with the largest effect sizes in the TWAS prediction model for LIME1 are in high LD with rs6062304, whereas those for ZGPAT are not.