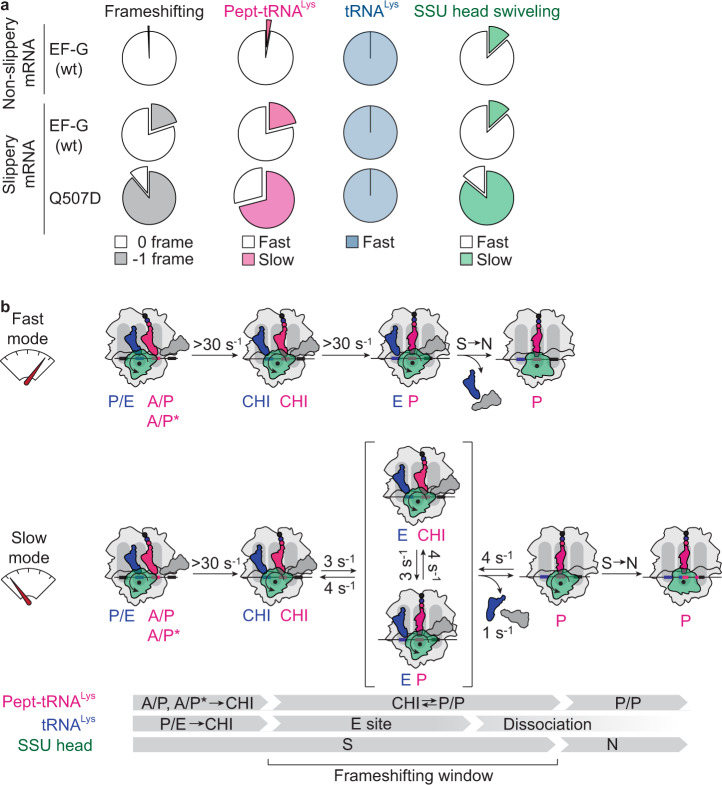
Fig. 7. Translocation trajectories correlating with frameshifting.
a Pie charts comparing the frameshifting efficiency (gray) with the distribution of translocation rates for pept-tRNA (magenta) and deacylated tRNA (blue) and SSU head domain back rotation (green) on non-slippery and slippery mRNA with EF-G(wt) and EF-G(Q507D). b Kinetic model of translocation on slippery mRNA. Majority of ribosomes translocate in fast mode with tRNAs moving synchronously to the POST state; back swiveling of the SSU head domain completes translocation. A fraction of ribosomes translocates in slow mode where pept-tRNA is trapped fluctuating between CHI and P/P instead of moving to the POST state. Deacylated tRNA translocates rapidly and dissociates from the E site allowing pept-tRNA to sample 0- and –1-frame codons. SSU head closure is delayed due to prolonged fluctuations of pept-tRNA. Rates of the elemental reactions for pept-tRNA are indicated.