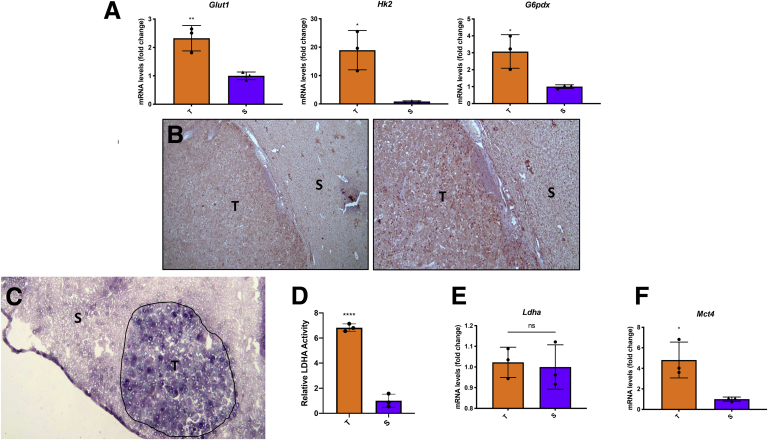
Figure 2.
(A) qRT-PCR analysis of Glut1, Hk2, and G6pdx in tumors compared with the surrounding areas. (B) Microphotographs showing G6pd immunostaining in HCC compared with peritumoral areas (G6pd staining: 5× [left], 10× [right]. (C) LDHA activity in tumors (T) and surrounding areas (S). (D) Relative LDHA activity. Measurement of LDHA activity was performed using the ImageJ software. (E) qRT-PCR analysis of Ldha. (F) qRT-PCR analysis of Mct4. qRT-PCR analysis was performed on dissected tumors or surrounding livers. Gene expression is reported as a fold change of tumor mRNA relative to surrounding livers. Relative quantification analysis for each gene was calculated by the 2ΔΔCt method. The histogram represents the means ± SD of 3 tumors and 3 surrounding livers (t test). ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001.