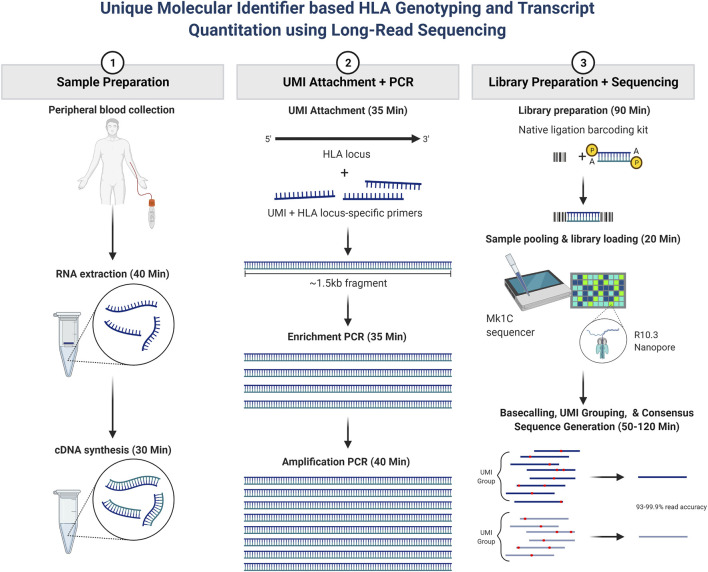
FIGURE 1.
Patient sample workflow for RNA isolation, library preparation, and resultant HLA typing as described based on consensus sequence generation. In short, total RNA was isolated from peripheral blood, and a portion of the total RNA was purified. RNA fragmentation and quantitation were determined, and an RNA integrity number (RIN) was calculated for each sample. cDNA synthesis was then performed, after which PCR amplification and purification were utilized to achieve the final enriched HLA-locus specific UMI tagged amplicon. The HLA gene-specific amplicons for each patient were then used for library preparation and bar coding with the Oxford Nanopore Technologies (ONT) platform. Sequencing was performed using the MinION R10.3 Flow Cell and loaded on the MinION Mk1C using the high-accuracy base calling module. The NGSengine bioinformatics pipeline was then utilized to provide HLA typing and Athlon2 was used to generate HLA allele-specific UMI counts.