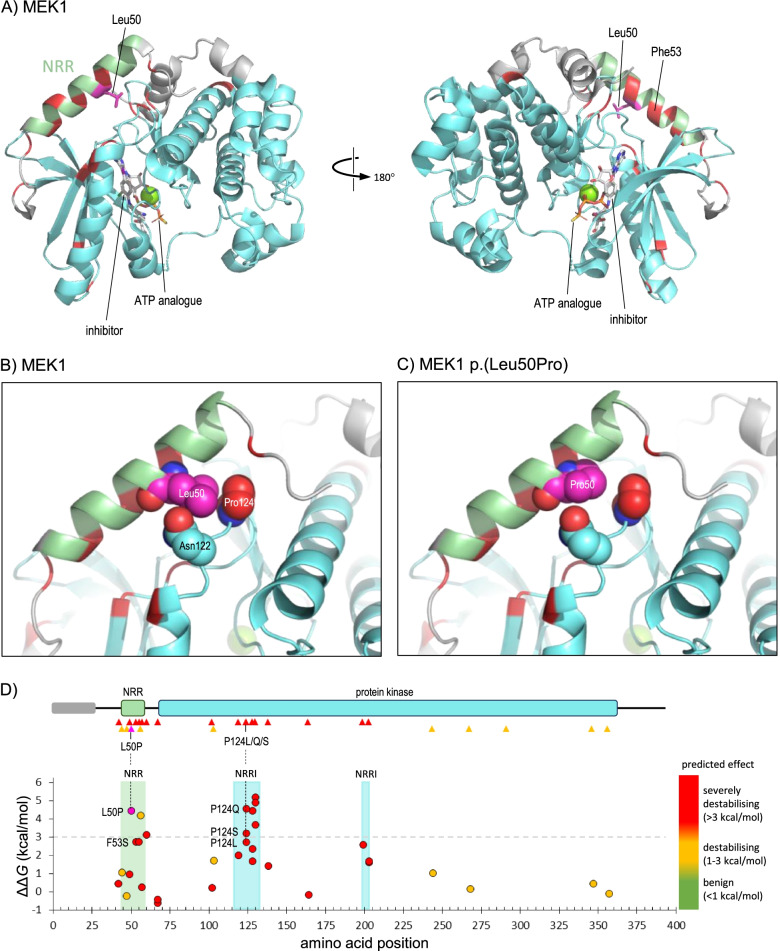
Fig. 4.
MAP2K1 NM_002755.4:c.149 T > C p.(Leu50Pro). A Structure of MEK1 residues 39–381 in complex with an adenosine triphosphate (ATP) analogue and an inhibitor compound (PDB 3eqc); default colouring is grey, with residues of the NRR (44–58) and kinase domain (68–361) coloured light green or cyan, respectively; additionally, Leu50 is coloured magenta, with the sidechain shown in stick format, while positions of missense variants reported as pathogenic in HGMD (class DM) are coloured red. B As A, but magnified to show detail around Leu50, for which all atoms are shown as space-filling spheres; spheres are also shown for sidechains atoms of Asn122 (carbon atoms cyan) and Pro124 (carbon atoms red), which lie in van der Waals contact with Leu50. C As B, but showing the predicted structure of the p.(Leu50Pro) variant. D The upper part shows the schematic organisation of MEK1; the grey bar indicates a region of predicted disorder (residues 1–27), while green and cyan bars show the NRR and protein kinase domains, respectively; triangles below show the location of variants reported in HGMD (red, pathogenic/class DM; orange, possibly pathogenic/class DM?), while the site of the p.(Leu50Pro) variant is shown by a magenta triangle. The lower part shows the predicted thermodynamic effect of the VUS p.(Leu50Pro) (magenta fill) and all HGMD missense variants (red fill, class DM; orange fill, class DM?) on MEK1 stability calculated in PDB 3uqc, and is aligned to the upper schematic; the light green-shaded region in the graph shows the extent of the NRR (green shading), while cyan-shaded regions show residues of the kinase domain which lie in contact with the NRR (NRRI: NRR-interacting); note that the most destabilising variants, including p.(Leu50Pro), all occur in the NRR or NRRI regions; these include three variants at Pro124, which interacts directly with Leu50 (vertical broken lines)