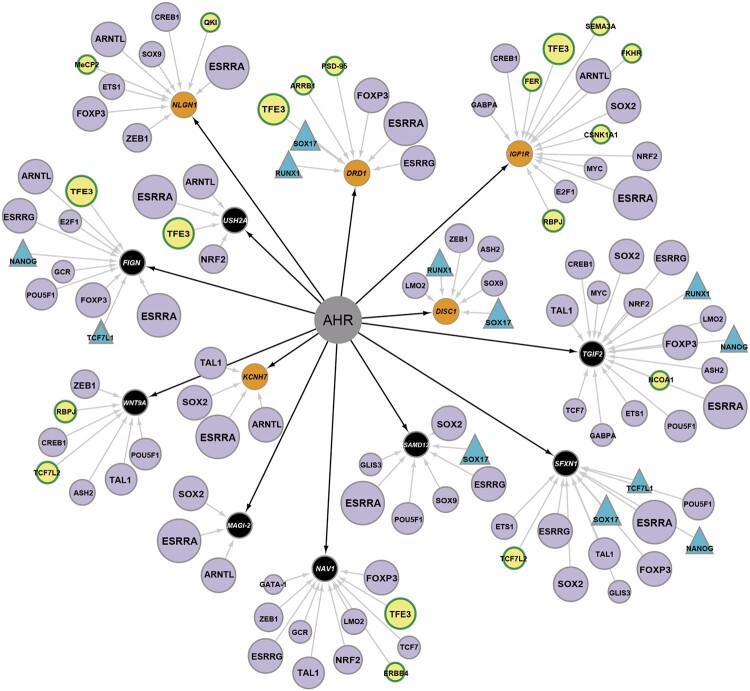
Figure 8.
Transcriptional regulation of aryl hydrocarbon receptor (AHR)-associated differentially expressed genes in wild-type and wfikkn1 mutant zebrafish exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Orange circles (NLGN1, DRD1, IGF1R, DISC1, KCNH7) depict the genes from Cluster 2 (Figure 6) and belong to at least one of the enriched biological process networks (Figure 7) that are predicted to have direct interactions with the AHR. Black circles (TGIF2, SFXN1, SAMD12, NAV1, MAGI-2, WNT9A, FIGN, USH2A) are genes in Cluster 2 that do not belong to a biological process network but have a direct interaction with the AHR. The transcription factors (TFs; purple) either belong to the top 30 predicted enriched TFs for the dataset or are found in the dataset (yellow). TF circle sizes refer to the number of predicted targets here, with the largest circles having 10 targets (eg, ESRRA), and the smallest circles having 1 target (eg, QK1 and FER). TFs are shown as triangles interact with wfikkn1 in MetaCore’s manually curated database. A color version of this figure appears in the online version of this article.