Figure.
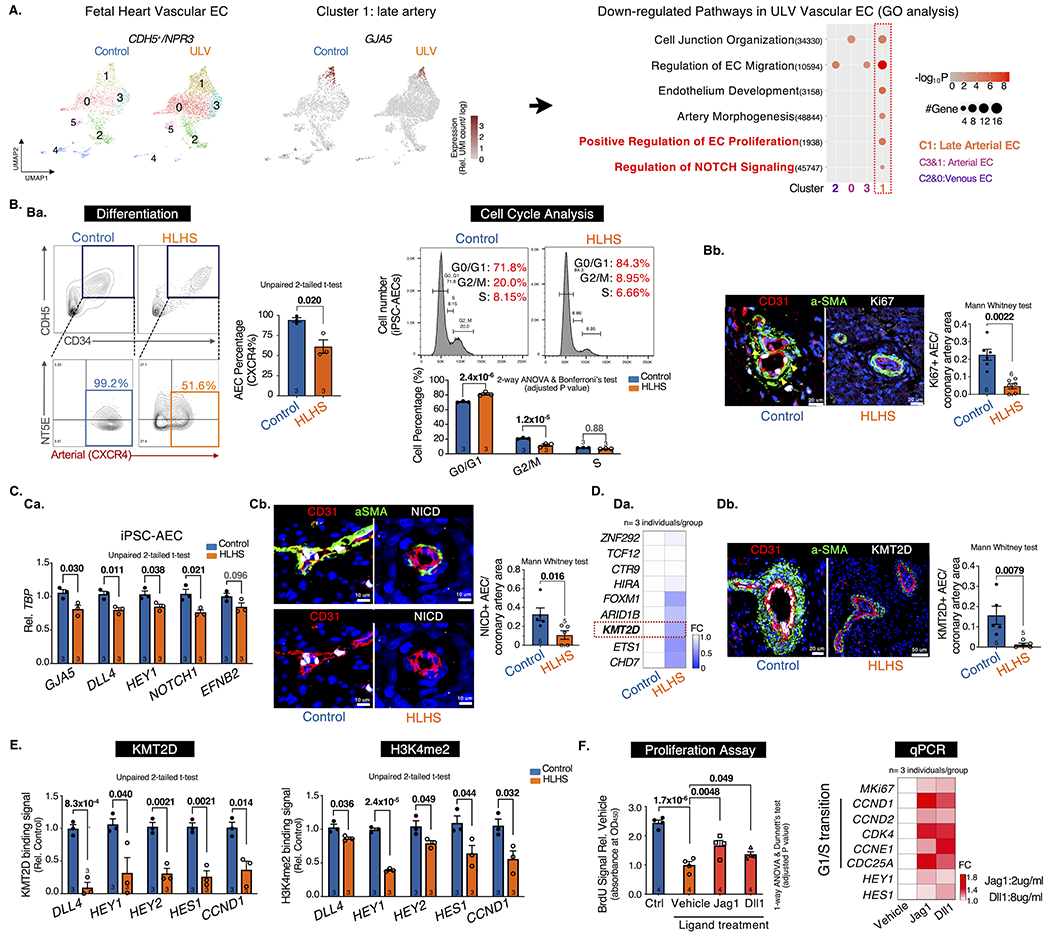
A. scRNA-seq analysis of vascular ECs (CDH5+/NPR3−). UMAP projection of Control vs. ULV human fetal left heart vascular ECs were shown on the left. Cluster1: GJA5+ late artery. Cluster4: venous-to-endocardial transitional population. N=1 per group. Then Gene Ontology analysis were performed on each cluster and showed downregulated pathways in ULV vascular ECs vs. control. GO terms: adjusted p-value < 0.05. UMI: Unique Molecular Identifier.
B. Artery differentiation and proliferation were impaired in HLHS. Ba, flow cytometry analysis of iPSC-AEC progenitors percentage (left) and their cell-cycle statuses (right). Bb, Immunostaining and quantification of Ki67+ arteries in human heart tissue. aSMA: smooth muscle actin.
C. NOTCH pathway was suppressed in HLHS AECs. Ca, qPCR of NOTCH genes in iPSC-AECs. Cb, Immunostaining and quantification of NICD+ arteries in human heart tissue.
D. KMT2D expression was decreased in HLHS AECs. Da, qPCR of HLHS DNM gene expression in iPSC-ECs. Db, Immunostaining and quantification of KMT2D+ arteries in human heart tissue.
E. KMT2D binding capacity and H3K4me2 signal were reduced in HLHS iPSC-AECs. (Evaluated via ChIP-qPCR)
F. NOTCH ligands treatment improved the proliferation of HLHS iPSC-AECs. Both BrdU proliferation assay and qPCR showed improved proliferation in HLHS iPSC-AECs. N=4 technical repeats. FC: fold change normalized to control.
Statistics (GraphPad Prism 9.3.1): Based on additional literature support from similar studies, our samples fit normal distribution. Parametric test: unpaired 2-tailed t-test (2 groups), ANOVA (>2 groups) with post hoc tests as indicated; non-parametric test: Mann-Whitney (2 groups). Mean±SEM; n= biological replicates as indicated.
