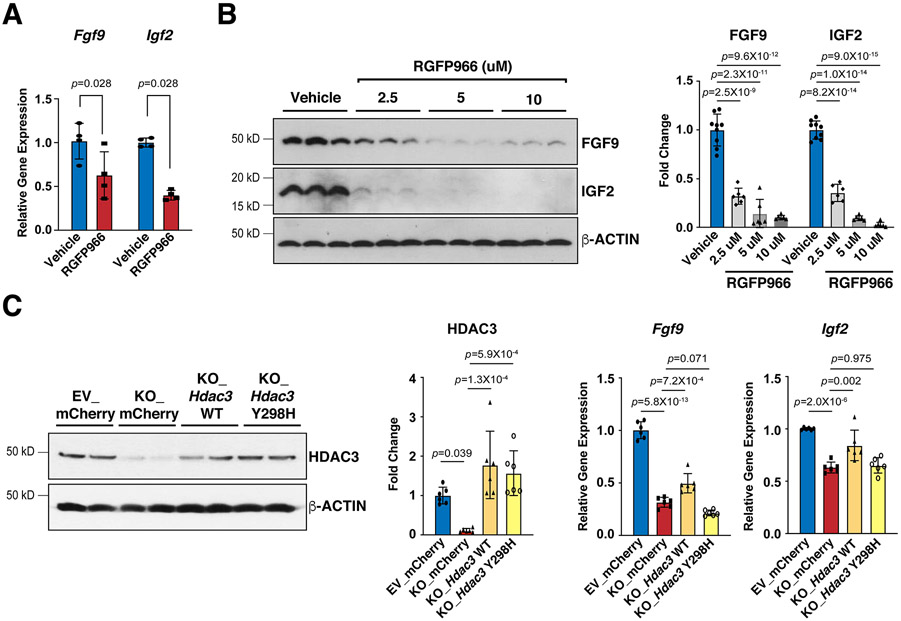
Figure 4. HDAC3 induces the expression of FGF9 and IGF2 dependent on its deacetylase activity.
(A) Decease of Fgf9 and Igf2 mRNAs after RGFP966 (a selective Hdac3 inhibitor) treatment. MECs were treated with 10 uM RGFP966 or vehicle for 24 hours. mRNA levels were quantified by qRT-PCR. Gapdh was used as a cDNA loading control. Fgf9, n=4 in each group; Igf2, n=5 in each group. (B) Decease of FGF9 and IGF2 protein levels after RGFP966 treatment. MECs were treated with 2.5 uM (n=6), 5 uM (n=6) or 10 uM (n=6) RGFP966 or vehicle (n=9) for 24 hours. FGF9 and IGF2 were quantified by western blot. GAPDH was used as a protein loading control. (C) mRNA levels of Fgf9 and Igf2 in Hdac3 KO and EV MECs after 24 hours treatment with Hdac3 WT (wild type), Y298H mutant, or mCherry (CTL) lentivirus. The expression of HDAC3 was quantified by western blot. n=6 in each group. The mRNA levels of Fgf9 and Igf2 were quantified by qRT-PCR. n=6 in each group. P-values were determined by the Mann-Whitney U test for (A) and One-way ANOVA followed by the Tukey post hoc test for (B) and (C).