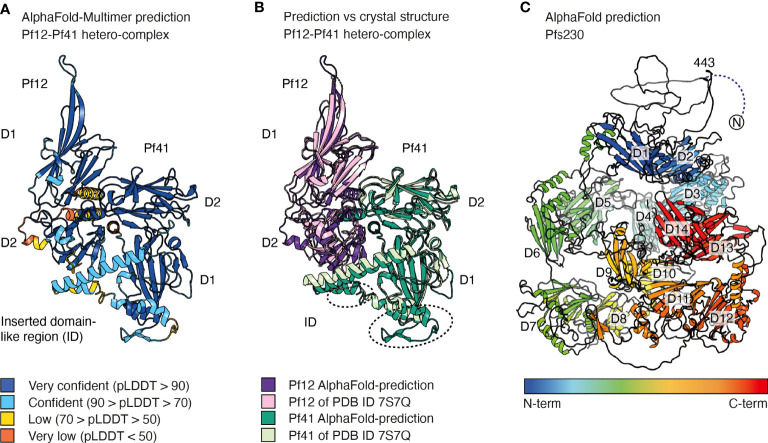
Figure 4.
AlphaFold predictions of selected 6-cysteine proteins. (A) AlphaFold-Multimer prediction of the hetero-dimeric complex of Pf12 and Pf41. Amino acids are colored based on their per-residue confidence score (pLDDT), which values can be between 0 and 100. Low values indicate low confidence and high numbers indicate very confident predictions. Amino acids are either colored orange (pDLLT <50), yellow (70> pDLLT >50), light blue (50< pDLLT >70), or dark blue (pDLLT >90). Regions with pLDDT <50 may be unstructured in isolation. (B) Alignment of the AlphaFold prediction (dark purple and dark green) of the Pf12-Pf41 hetero-dimeric complex with the Pf12-Pf41 crystal structure (light purple and light green, PDB ID 7S7Q). Regions of the Pf41 ID which are not defined in the crystal structure are indicated by dotted circles. (C) AlphaFold prediction of Pfs230 colored from blue to red from N- to C-terminus. The N-terminal residues 1-575 have low pDLLT values of <50. Residues 1-442 are not shown for clarity of the image. The 14 6-cysteine domains are indicated from D1-D14. AlphaFold models were generated at WEHI (Australia) using the AlphaFold algorithm.