Fig. 4 |. Allele-specific knockdown of transcripts containing SPTLC1 variants associated with ALS.
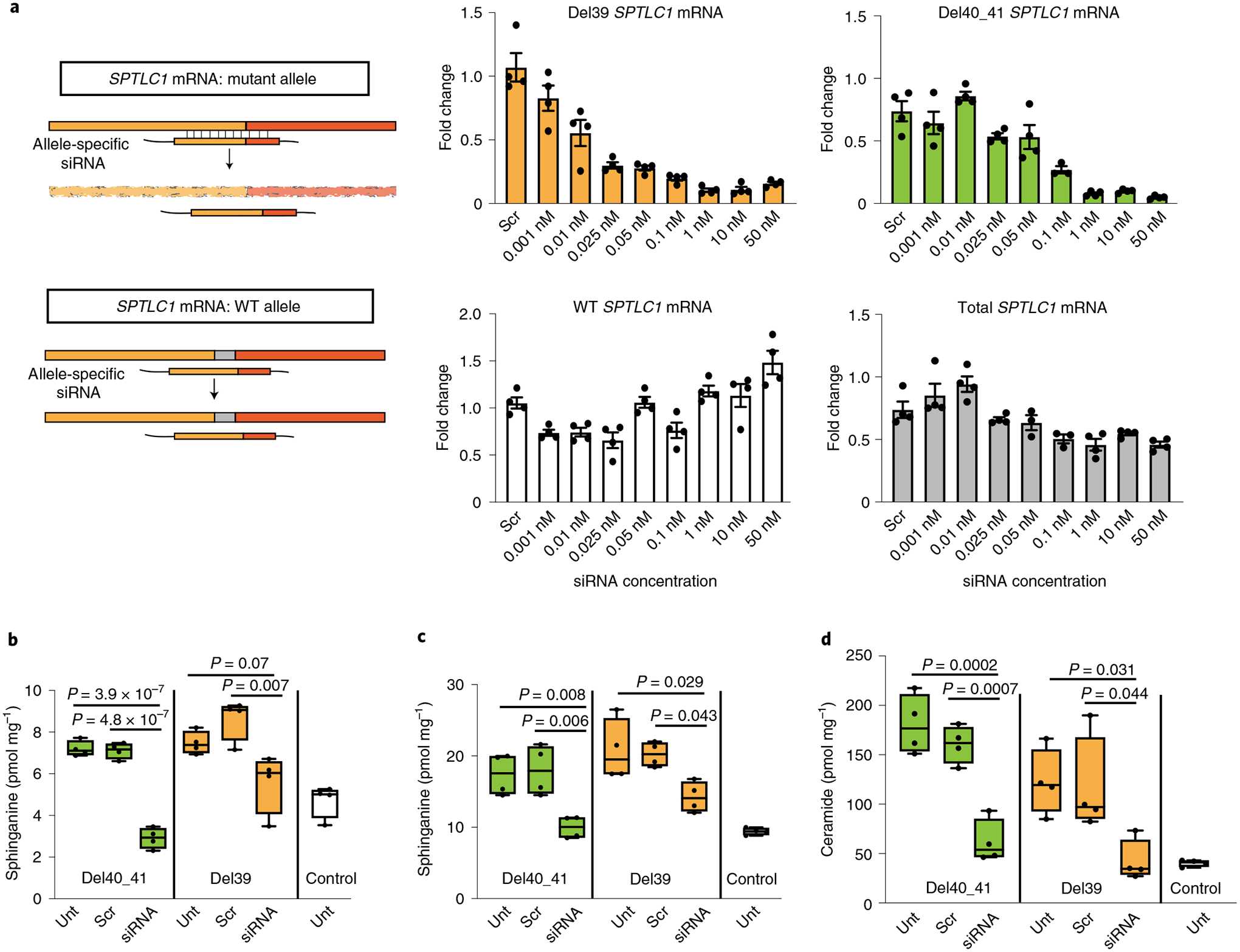
a, Allele-specific knockdown using siRNA. Precision-designed, mutation-specific siRNAs target the variant allele mRNA while not interfering with the WT transcript. Bar graphs show real-time reverse transcriptase PCR results from patient-derived fibroblasts treated with varying concentrations of allele-specific siRNAs. All values are reported as linear fold change compared to untreated cells in the same experiment. The levels of mutant allele transcripts L39del (yellow) and F40_S41del (green) show a dose-dependent decrease with increasing siRNA concentration. The levels of WT allele transcript in L39del cells (white) show no notable change, indicating allele specificity. In F40_S41del cells (gray), the level of total transcript including both mutant and WT alleles was measured and did not decrease by more than the expected 50% of total, akin to haploinsufficiency. Error bars are s.e.m. from four replicates. b–d, Corrective effect of allele-specific siRNA treatment on de novo synthesis of sphinganine (b), sphingosine (c) and ceramide (d) levels in patient-derived fibroblasts. Cells were treated with siRNA and labeled with 3,3-D2 l-serine (3 mM) 24 h before analysis. Data represented by box-and-whisker plots are the median, first and third quartiles, and minimum and maximum from four replicates. Ordinary one-way ANOVA with Tukey’s adjustment for multiple comparisons was used for statistical comparisons. Unt, untreated; scr, scrambled.
