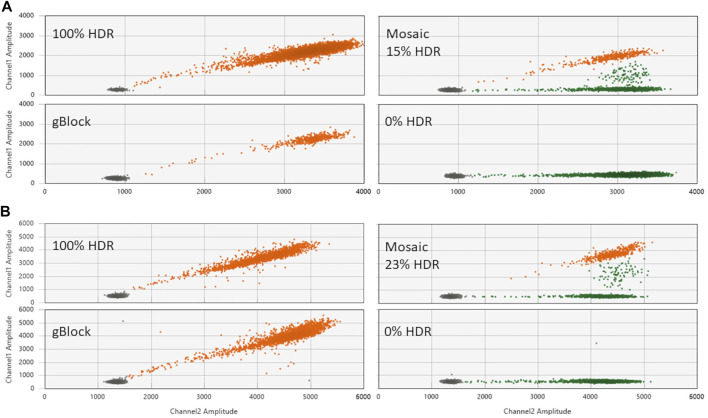
FIGURE 2.
Evaluation of the contribution of intended PMEL and PRLR mutations in edited embryos by ddPCR. (A) Shown are 2D amplification plots generated with three different embryos that were injected with plasmid encoded TALENs TL17/TR217 plus ssODN 1288 and analyzed with the HDR probe 1321 for the PMEL ΔCTT (p.Leu18del) mutation. The panel labeled as gBlock depicts the assay results for a synthetic DNA fragment (1282) comprising the PMEL ΔCTT (p.Leu18del) mutation and represents a positive control for 100% HDR. (B) ddPCR results for three different embryos injected with plasmid encoded gRNA 632/Cas9 plus ssODN 1403 and analyzed with the HDR probe 1554 for the PRLR T > A (p.C440*) mutation. The 100% HDR control panel (gBlock) was done with synthetic DNA fragment 1413. Orange dots represent HDR-positive droplets, green droplets represent wt and non-HDR indel alleles and black dots represent droplets with no amplification. Contributions of the intended HDR alleles are given for each plot with examples for embryos that were fully converted to the HDR genotype (100% HDR), only partially HDR edited embryos (Mosaic) and embryos without edited HDR alleles (0% HDR). Channel1 Amplitude: FAM signal (HDR probe); Channel2 Amplitude: HEX signal (reference probe).