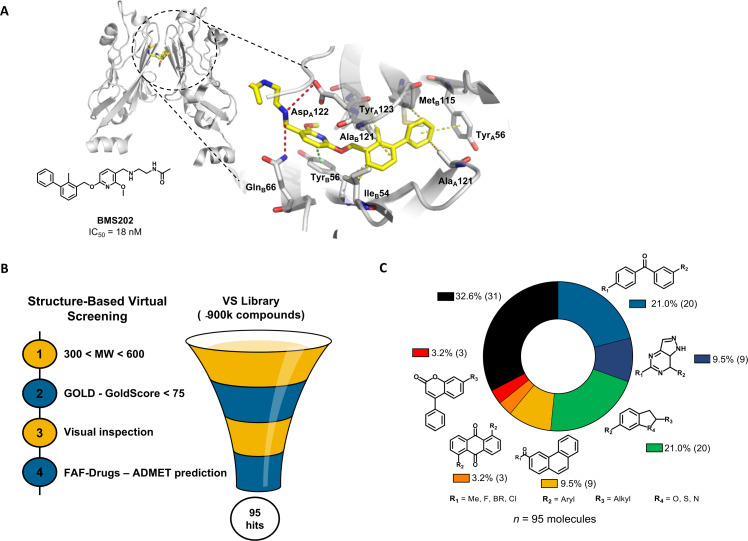
Figure 1.
In silico virtual screening for putative PD-1/PD-L1 inhibitors. (A) Ribbon representation of PD-L1 monomers (gray) bridged by small-molecule inhibitor BMS202 (yellow) (PDB 5J89). Close-up view of the binding pocket. Receptor-ligand interactions are displayed in dashes. Hydrophobic contacts (yellow), π-staking (green) and H-bond and salt bridges (red). (B) Structure-based virtual screening for the identification of PD-1/PD-L1 small-molecule inhibitors, beginning with pre-filtering the compounds based on molecular weight (MW), followed by the screening in silico using the scoring function GoldScore of the GOLD software, followed by the visual inspection of the top-ranked compounds within the binding pocket. Finally, the compounds were uploaded into the FAF-Drugs predictor to address the administration, distribution, metabolism, excretion, and toxicity (ADMET) properties. (C) In total, 95 chemically diverse virtual hits were selected to move forward. GOLD, Genetic Optimization for Ligand Docking; PD-1, programmed cell death protein 1; PD-L1, PD-ligand 1; vs, virtual screening