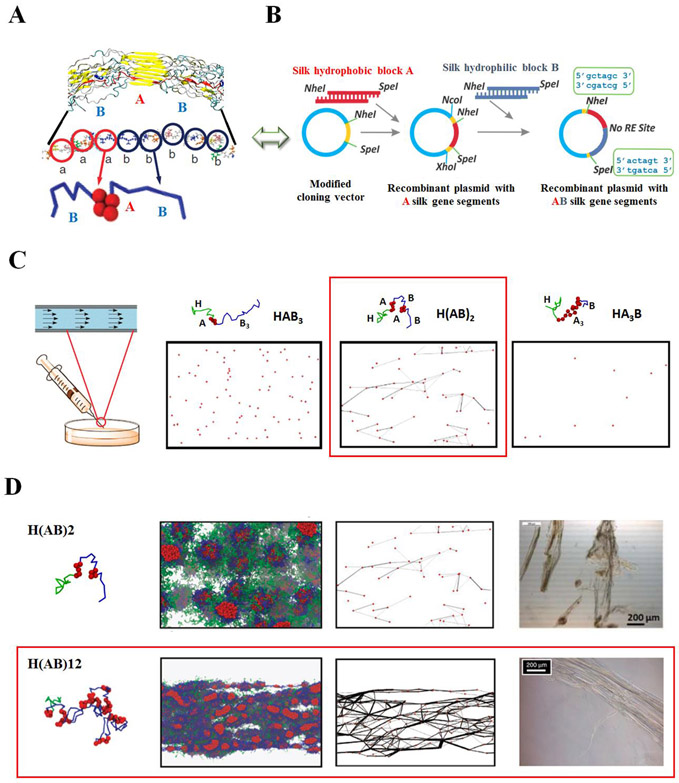
Figure 3.
Integrated modeling and experimental approaches for the production of robust recombinant spider silk fibers. Schematic of (A) coarse-graining simulation, (B) recombinant DNA strategy and (C) processing method (e.g., wet spinning) used to study spider silk block copolymers. (C) Coarse-grained representations, and the mesoscopic dissipative particle dynamics (DPD) simulation node-bridge diagram under the shear flow of the recombinant spider silk peptides: HAB3, H(AB)2 and HA3B. Simulation suggested that under shear flow spinning process, only the H(AB)2 sequence (highlighted in red box) with alternating hydrophobic/hydrophilic domains at a 1:1 ratio promote network formation over other domain distributions. (D) Coarse-grained representations, mesoscopic DPD simulation snapshots and the node-bridge diagram under shear flow, and the bright-field microscopic images of spun fibers in bundles in the coagulation bath of the recombinant spider silk peptides: H(AB)2 and H(AB)12. As length is increased from H(AB)2 to H(AB)12, mature networks for robust spider silk fibers formed in H(AB)12 (highlighted in red box) under shear flow. Red beads - hydrophobic ‘A’ beads in the ‘A’ domain, blue lines - hydrophilic ‘B’ beads in the ‘B’ domain and green lines - hydrophilic ‘B’ beads in the ‘H’ domain. Water beads are not shown for clarity. Adapted with permission from ref. 11 and 38. Copyright 2014 Elsevier Inc., and 2015 Nature Publishing Group.