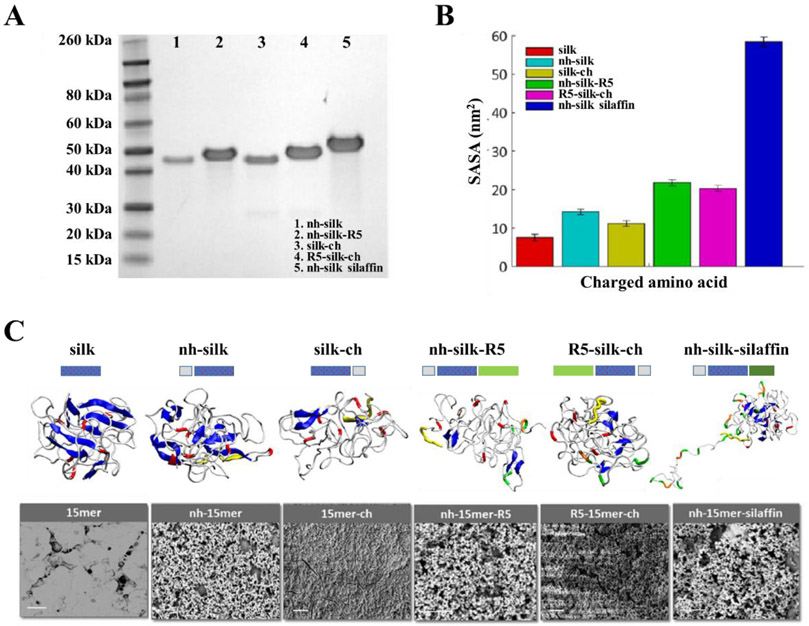
Figure 4.
Integrated modeling and experimental approach for the design of biomineralization interfaces. (A) SDS-PAGE of recombinant silk-silica fusion proteins: nh-silk (40 kDa), nh-silk-R5 (43 kDa), silk-ch (40 kDa), R5-silk-ch (43 kDa) and nh-silk-sillafin (55 kDa). (B) Solvent accessible surface area (SASA) of positively charged amino acids in recombinant silk-silica fusion proteins. (C) Schematics of the silk-silica fusion protein designs, REMD simulation of protein folding, and SEM evaluation of the ability to induce silica formation. Correlation between REMD snapshot and SEM images suggested that the folding and exposure of charged functional groups played key roles in biomineralization. Peptide design: his-tag - grey box, spider silk polymer – dotted blue box, and R5 domain - green box. REMD snapshot: β-sheets – blue, random coils - white, histidine - yellow, arginine (in silk domain) - red, arginine (in R5 binding peptide) - orange, and lysine - green. SEM image scale bars are 10 μm. Adapted with permission from ref. 26. Copyright 2016 American Chemical Society.