FIGURE 1.
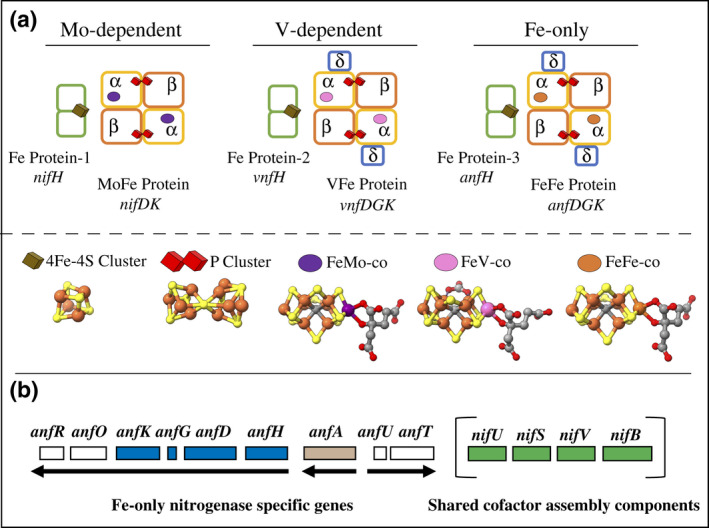
Schematic representation of the catalytic components of the three nitrogenases found in Azotobacter vinelandii. (a) the MoFe protein is a tetramer that contains two pairs of metalloclusters: P‐cluster located at the interface of the α‐β subunits, and FeMo‐cofactor located within the α‐subunits. VFe‐ and the FeFe‐nitrogenase architectures are like the Mo‐nitrogenase, except for the presence of a third subunit, δ, encoded by vnfG and anfG genes, respectively. Protein designations and their encoding genes are indicated below each component. The corresponding Fe proteins for the three systems are indicated as 1, 2 or 3. Atoms in the structures are indicated as follows: yellow, sulfur; gray: carbon; red: oxygen; orange: iron; purple, molybdenum; pink, vanadium. The coordinates for the different clusters represented were extracted from the following PDB files: [4Fe‐4S] cluster from PDB 2NIP; P‐cluster from PDB 3U7Q; FeMo‐cofactor and FeV‐cofactor from PDB 3U7Q and PDB 5N6Y, respectively. FeFe‐cofactor has been inferred from FeMo‐cofactor with Fe replacing Mo although its structure has not yet been established by crystallographic methods. Structures were visualized with ChimeraX software (Pettersen et al., 2020). (b) Schematic representation of the genes associated with the Fe‐only nitrogenase in A. vinelandii. Four nif gene products are necessary for the maturation of the FeFe protein (bracketed, in green). Nine genes are specifically associated with the Fe‐only system, designated as anf. The genes encoding the catalytic components (anfHDGK) are colored in blue; anfA, which encodes for the transcriptional activator, is colored in brown
