Figure 3.
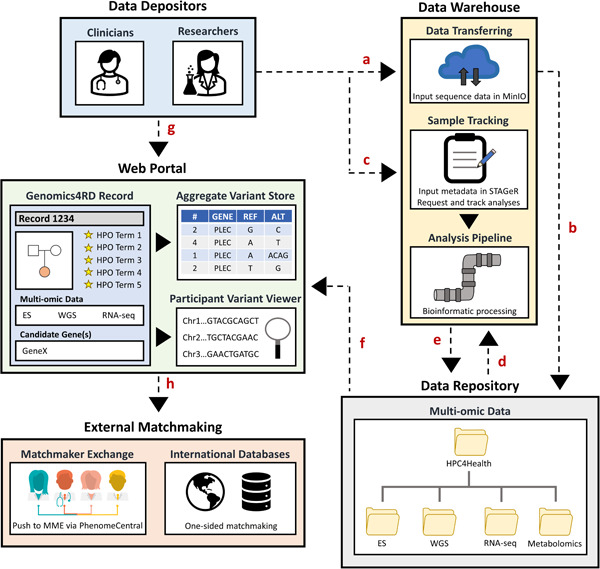
Genomics4RD data input and visualization. (a) Raw multiomic sequencing files are uploaded into the secure MinIO server for (b) deposition into the data repository. (c) Metadata about participants are entered into a laboratory management system called STAGeR. (d) Raw multiomic data files are run through their respective bioinformatic pipelines for processing and (e) stored in the Genomics4RD data repository. (f) The catalog of available multiomic data sets will be displayed for each participant record, and the filtered genomic results are directly browsable through the variant viewer interfaces. (g) Data depositors may access the Genomics4RD user interface via the web portal, where they complete coded records for their participants (including inputting clinical data, pedigrees, prioritized HPO terms, and candidate genes). Within the web portal, users can visualize their participants' processed files via the participant variant viewer and query phenotypic and variant‐level data across Genomics4RD. (h) Further matchmaking opportunities can be explored by pushing Genomics4RD records to the PhenomeCentral node of the Matchmaker Exchange (for two‐sided matchmaking), and through connections with other international databases (for one‐sided matchmaking). HPO, human phenotype ontology
