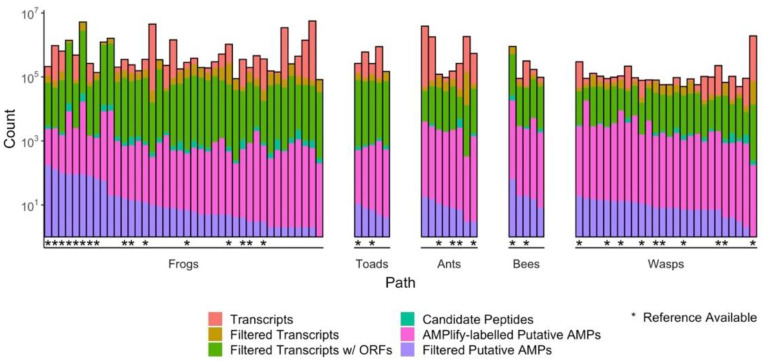
Figure 1.
Statistics and attrition as the sequencing data are processed by the rAMPage AMP discovery pipeline. rAMPage processes RNA-seq datasets from raw reads to transcripts to putative AMPs. In this case, a putative AMP is defined as a sequence with an AMPlify score 10 for amphibians or 7 for insects, a length 30 AA, and a charge 2. Datasets with a reference transcriptome used during assembly are indicated with an asterisk. The total number of putative AMPs (n = 1478, including 341 duplicates) are shown in purple, discovered from a total of ~53 million assembled transcripts.