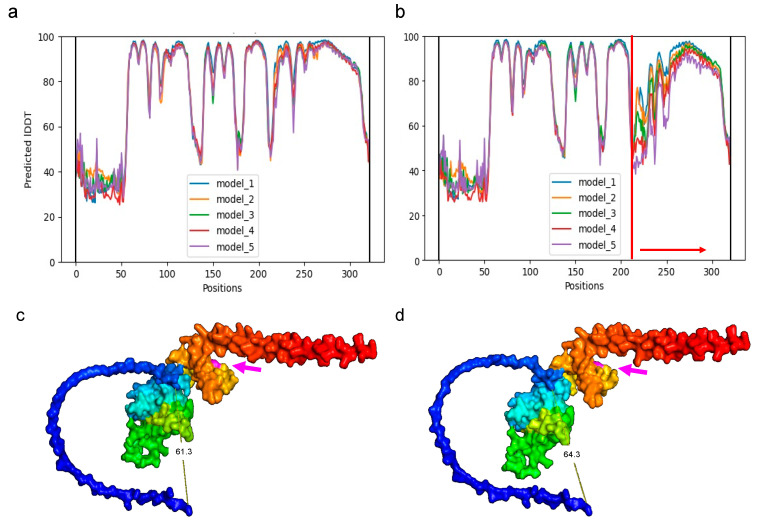
Figure 6.
(a) Predicted local Distance Difference Test (lDDT) for the PURA WT and (b) PURA–Phe233del-predicted structure deletion per amino acid using five different models via AlpahFold2. Models show that the occurrence of the Phe233del affects between 220–320 amino acids (red arrow). (c) A 3D reconstruction of the transcriptional activator protein Pur-alpha (PURA; Uniprot: Q00577) from the WT sequence using AlphaFold, focusing on the surrounding area of the Phe233 amino acid (in magenta; magenta arrow). (d) A 3D reconstruction of the PURA–Phe233del. Note the distortion (61.3 Å vs. 64.3 Å) in PURA structural conformation after the p.Phe233del occurs.