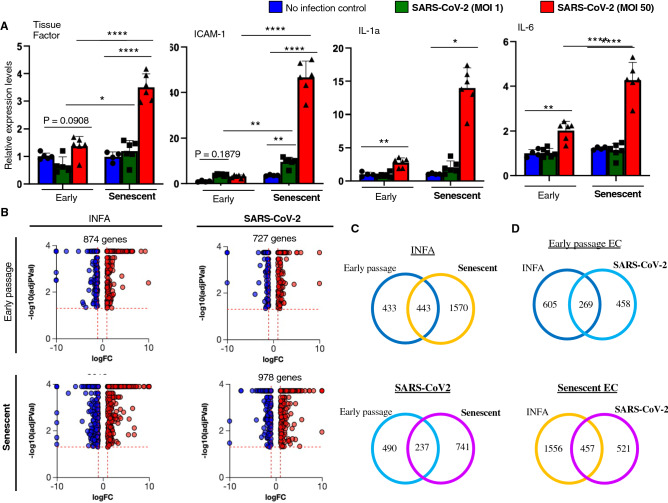
Figure 3.
SARS-CoV-2 infection affects various genes expression in ECs. (A) Quantitative PCR analysis for tissue factor (TF), intercellular adhesion molecule-1 (ICAM-1), interleukin-1a (IL-1a), and IL-6 in early passage and senescent HUVECs with or without SARS-CoV-2 infection (1 or 50 MOI) at 72 hpi (n = 4 each for control cells; n = 6 each for infected cells). Statistical analyses were performed using one-way ANOVA with Fisher’s LSD post hoc analysis for TF, ICAM-1, and IL-6, while Kruskal-Wallis test by ranks was used for IL-1a. Data are presented as mean ± SE *P < 0.05, **P < 0.01, and ****P < 0.0001. (B) Comprehensive RNA-Seq data analysis in ECs infected with SARS-CoV-2 or INFA was performed. Volcano plots for genes whose expression was significantly altered (P < 0.05) by SARS-CoV-2 or INFA infection in early passage and senescent ECs were shown (n = 3 each). Red plots indicate the genes significantly increased more than 2-fold after the virus infection. Blue plots indicate the genes significantly (P < 0.05) decreased less than half after the virus infection. (C,D) Venn Diagram analysis of the genes whose expression was significantly altered by SARS-CoV-2 or INFA infection (> 2 or < 1/2 fold) in young and senescent ECs. Numbers indicate the number of genes included in the sets of the intersections.