Figure 3.
NK cell subsets associated with longitudinal trajectory of intact proviruses
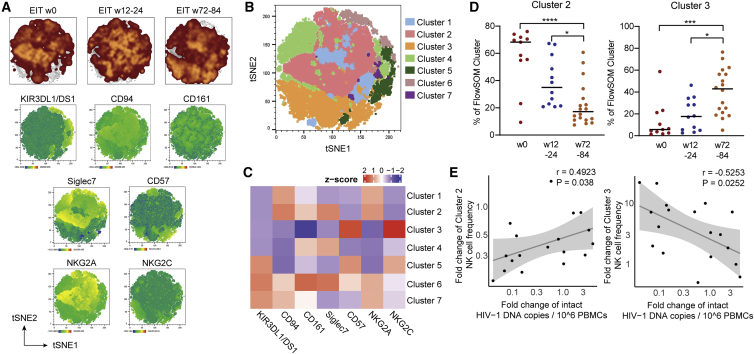
(A) (Top) Global t-distributed stochastic neighbor embedding (tSNE) maps of CD56dim CD16dim NK cells from early-treated infants; data from indicated time points are shown separately as overlays. (Bottom) tSNE maps showing the expression of individual phenotypic markers measured by flow cytometry in concatenated CD56dim CD16dim NK cells analyzed from early-treated infants.
(B) tSNE map displaying 7 phenotypically distinct clusters identified by FlowSOM within concatenated CD56dim CD16dim NK cells from early-treated infants.
(C) Heatmap showing the mean fluorescence intensity (MFI) of the phenotypic parameters measured in the 7 clusters shown in (B).
(D) Longitudinal evolution of clusters 2 and 3 NK cell populations from weeks 0 (n = 11), 12/24 (n = 12), and 72/84 (n = 18). ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; Kruskal Wallis test with post-hoc Dunn’s test.
(E) Correlation between longitudinal fold changes in proportions of cluster 2 or 3 NK cells (between weeks 0 and 72/84) and corresponding changes in intact proviruses from early-treated infants. Spearman correlation coefficient is shown.