Abstract
Fecal microbiota transplantation (FMT) has been reported to decrease the incidence of recurrent urinary tract infections (UTIs), presumably by restoring microbiome diversity and/or uropathogen competition. We report a 16-year-old female with recurrent UTIs caused by multidrug-resistant Klebsiella pneumoniae, for which frequent intravenous broad-spectrum antibiotic treatment was necessary. The patient was treated with FMT from a well-screened healthy donor without multidrug-resistant bacteria in the feces. After FMT, she developed several UTIs with an antibiotic-susceptible Escherichia coli that could be treated orally. The uropathogenic E. coli could be cultured from donor feces, and whole genome sequencing confirmed donor-to-recipient transmission. Our observation should stimulate discussion on long-term follow-up of all infections after FMT and donor fecal screening for antibiotic-susceptible Enterobacterales.
Keywords: fecal microbiota transplantation, FMT, MDRO, ESBL, uropathogen, urinary tract infection
Fecal microbiota transplantation (FMT) is recommended for patients with multiple recurrent Clostridioides difficile infections. In these patients, FMT also seems to decrease the load of antimicrobial resistance genes and the phylum Proteobacteria (which includes Enterobacterales) [1]. FMT has been explored for gut decolonization in patients with colonization and/or infections with multidrug-resistant organisms (MDROs). However, success rates for decolonization are heterogeneous while spontaneous decolonization has also been described [2, 3]. Recently, gut microbiome dysbiosis has been linked to recurrent urinary tract infections (UTIs) [4]. Several case reports and an observational study suggest that FMT may be an effective treatment to prevent recurrent UTIs [5–12].
Although FMT is generally considered safe, severe adverse events with transmission of multidrug-resistant Escherichia coli [13] and Shiga toxin–producing E. coli have been reported [14]. Consequently, the US Food and Drug Administration (FDA) issued safety warnings and recommends enhanced screening of donor stool. Here, we report a pediatric patient who underwent FMT because of recurrent MDRO UTIs and highlight transfer of a uropathogenic E. coli causing UTIs in the recipient.
CASE REPORT
An FMT was requested for a 16-year-old female with recurrent febrile UTIs and gut colonization with a multidrug-resistant (MDR) Klebsiella pneumoniae: extended-spectrum β-lactamase (ESBL)–producing, susceptible to fosfomycin, colistin, and meropenem, with variable susceptibility to nitrofurantoin. The medical history included familial holoprosencephaly, epilepsy, correction of scoliosis due to spasticity, cystic renal dysplasia, and feeding problems for which she had a percutaneous endoscopic gastrostomy tube. The past 2 years she had been regularly admitted for intravenous (IV) meropenem treatment for, on average, 1 UTI every 1–2 months. Other treatments included oral fosfomycin and intravesical gentamicin administration, yet without sustained response. The MDR K. pneumoniae was repeatedly isolated from urine, perineal swabs, and feces. While urine culture was negative directly following meropenem, fecal cultures were positive for K. pneumoniae, suggesting bacterial translocation from the gut via ascension in the urinary tract as underlying mechanism for the recurrent UTIs. However, the patient also had dysfunctional voiding as a possible contributing factor to recurrences, due to severe psychomotor retardation. No signs of focal infection were demonstrated on repeated ultrasound of the kidneys.
The UTIs led to renal scarring documented by DMSA (dimercaptosuccinic acid) scan. Multiple prolonged admissions for IV antibiotic therapy had a profound impact on the quality of life of the patient and her family. Two courses of meropenem within 1 month prompted a request for FMT via the compassionate use program of the Netherlands Donor Feces Bank (NDFB) to attempt gut decolonization and/or decrease the frequency of recurrent UTIs with the MDR K. pneumoniae. With no viable alternative treatment option left, the multidisciplinary NDFB working group deemed the patient eligible for FMT. Informed consent was obtained from the parents.
The patient received the FMT (198 mL, prepared from 60 g of feces) via an endoscopically placed duodenal tube under general anesthesia. Prior to FMT, a gram-negative gut decolonization scheme with polymyxin/neomycin 500 000 international units/125 mg orally 4 times daily combined with nitrofurantoin 100 mg orally twice daily was given for 4 days and stopped 24 hours pre-FMT. One day prior to FMT, 2 L of macrogol/electrolytes was administered via the percutaneous endoscopic gastrostomy tube. No complications occurred during the FMT procedure.
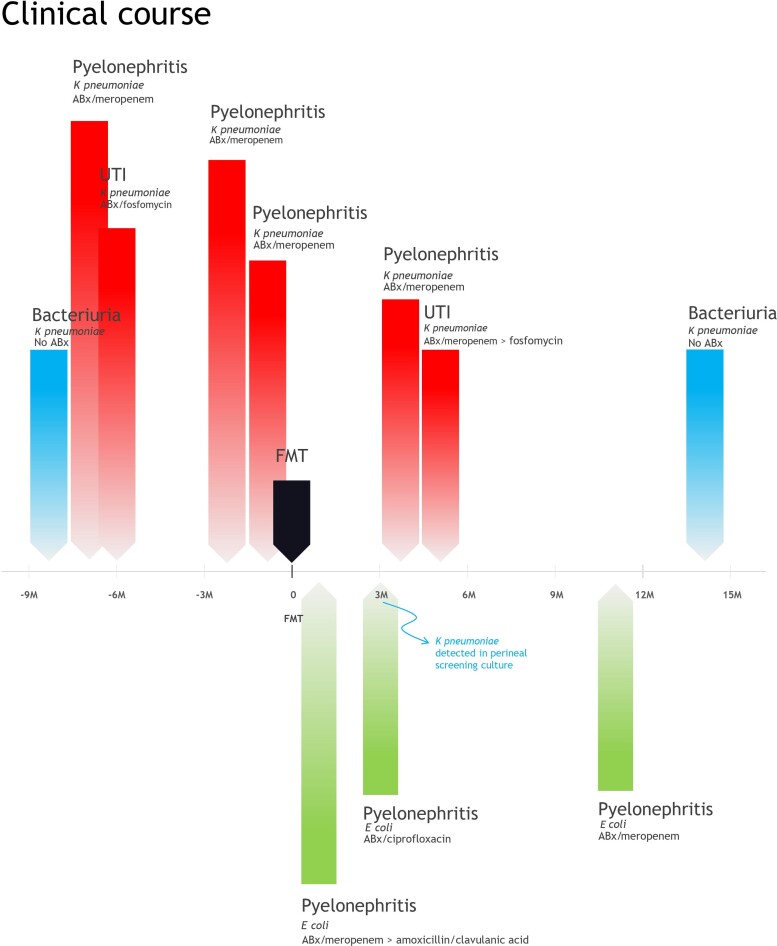
The clinical course is summarized in Figure 1. One month post-FMT, the patient was admitted with pyelonephritis caused by an amoxicillin/clavulanic acid–susceptible E. coli. After empiric meropenem treatment for 3 days (pending urinary culture results), she was discharged with amoxicillin/clavulanic acid orally (7 days). Three months post-FMT, a second episode of E. coli pyelonephritis was treated ambulatory with ciprofloxacin (14 days), though a perineal swab revealed return of the MDR K. pneumoniae. At 4 months, a Klebsiella pyelonephritis required IV meropenem (10 days). At 5 months, a suspected UTI was treated empirically with IV meropenem (1 day), but this was switched to fosfomycin (10 days) when deemed uncomplicated. At 11 months, pyelonephritis due to a ciprofloxacin- and amoxicillin/clavulanic acid–resistant E. coli was treated with IV meropenem. At 14 months, asymptomatic bacteriuria with the MDR K. pneumoniae was not treated.
Figure 1.
Clinical course of a patient who underwent fecal microbiota transplantation (FMT) for recurrent urinary tract infections (UTIs). Red bars indicate UTIs with positive urine cultures with multidrug-resistant (MDR) Klebsiella pneumoniae for which antibiotic treatment was given, blue bars indicate positive urine cultures with MDR K. pneumoniae that were not treated, and green bars indicate positive urine cultures with Escherichia coli that were treated. Abbreviations: ABx, antibiotic treatment; FMT, fecal microbiota transplantation; M, months relative to transplant; UTI, urinary tract infection.
MICROBIOLOGICAL ANALYSIS
We hypothesized that the E. coli associated with 3 UTI episodes post-FMT had been transmitted via donor feces. Feces from donor and patient were examined for the presence of antibiotic-susceptible and MDR Enterobacterales. Of the patient, 2 pre-FMT fecal samples were available: before (day FMT –4) and after (day FMT –1) antibiotic pretreatment, and 3 samples after FMT: 1, 8, and 17 months post-FMT. In addition, 2 different E. coli isolates cultured from a urine sample 3 weeks post-FMT were available: 1 ciprofloxacin-susceptible and 1 ciprofloxacin-resistant E. coli (minimum inhibitory concentration [MIC] ≤.25 mg/L and 1 mg/L, respectively). Both E. coli isolates were resistant to trimethoprim-sulfamethoxazole (TMP-SMX) (MIC >320 mg/L) and susceptible to amoxicillin/clavulanic acid (MIC ≤2 mg/L).
Culture
Raw feces aliquots were stored at −80°C. After thawing at room temperature, 10 µL was cultured with enrichment broth with subsequent plating on growth media, as previously described [15]. Colonies morphologically suspect for Enterobacterales were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Microflex, Bruker Daltonik, Bremen, Germany). Susceptibility testing was performed with the VITEK2 system (bioMérieux; Marcy-l'Étoile, France) using European Committee on Antimicrobial Susceptibility Testing breakpoints. ESBL production was confirmed using the double disk method.
The donor feces contained an E. coli with a similar antimicrobial resistance pattern as the E. coli detected in the patient’s urine, resistant to TMP-SMX (MIC ≥320 mg/L) with a ciprofloxacin MIC of 0.5 mg/L. Additionally, a TMP-SMX–susceptible E. coli, a non-MDR K. pneumoniae, and Enterobacter cloacae were cultured from the donor feces.
In the pre-FMT feces sample of the patient before antibiotic pretreatment (day FMT –4), the MDR K. pneumoniae was detected, in contrast to the sample after antibiotic pretreatment (day FMT –1). The MDR K. pneumoniae was again cultured from feces 1 month and 17 months post-FMT, but not at 8 months. The pre-FMT fecal sample and all post-FMT fecal samples of the patient were negative for the presence of the antibiotic-susceptible E. coli.
Whole Genome Sequencing
Whole genome sequencing (WGS) of E. coli isolates from the donor feces (n = 2, D1 and D2) and clinical patient urine sample (n = 2, P1 and P2) was performed to assess the relatedness between the strains and the presence of urovirulence factors and antibiotic resistance genes. DNA isolation and sequencing were performed as previously described [15].
The 4 E. coli isolates belonged to multilocus sequence typing (MLST) sequence type 69 and differed with a maximum of 4 alleles based on an in-house whole genome MLST scheme (comprising 4503 genes) of the Dutch National Institute for Public Health and the Environment (RIVM) [16]. This indicates clonal relationship since the cluster cutoff was established at ≤25 alleles. Single-nucleotide polymorphism (SNP) analysis using CLC Genomics workbench version 22 resulted in a maximum difference of 6 SNPs. The isolates formed a separate cluster when compared with MDR E. coli isolates from the national database of the RIVM, and belonged to Clermont phylotype D, which is associated with uropathogenicity [17]. Thirteen putative urovirulence factors (PUFs) and 8 additional urovirulence factors were identified (see Supplementary Table 1). Results of genomic antibiotic resistance analysis with the ResFinder database are shown in Table 1.
Table 1.
Antimicrobial Resistance Genes Identified in Escherichia coli Isolates From Donor Feces and Patient Urine
| Donor Escherichia coli | Patient Escherichia coli | |||
|---|---|---|---|---|
| Antimicrobial Resistance Gene | D1 | D2 | P1 | P2 |
| aadA5 | X | X | X | |
| dfrA17 | X | X | X | |
| mdf(A) | X | X | X | X |
| mph(A) | X | X | X | |
| gacE | X | X | X | |
| qnrB19 | X | X | X | X |
| sitABCD | X | X | X | X |
| sul1 | X | X | X | |
In 1 donor isolate (D1), only fluoroquinolone resistance (qnrB19), multidrug efflux pump (mdf(A)), and hydroxide peroxide resistance (sitABCD) genes were detected. These genes were also detected in donor isolate D2 and patient isolates P1 and P2, with additional presence of genes for antiseptic (gacE), macrolides (mph(A)), aminoglycosides (aadA5), trimethoprim (dfrA17), and sulfonamide (sul1) resistance. Abbreviation: X, gene present.
In addition, WGS was performed on K. pneumoniae isolates cultured from patient feces pre-FMT (P3) and 1 month post-FMT (P4), and clinical K. pneumoniae isolates cultured from urine pre-FMT (P5 and P6) and from a perineal swab 3 months post-FMT (P7). Core genome MLST analysis with Ridom SeqSphere+ indicated clonal relationship of the K. pneumoniae isolates: all isolates belonged to sequence type 307 with a maximum number of 5 alleles’ difference.
Microbiota Analysis
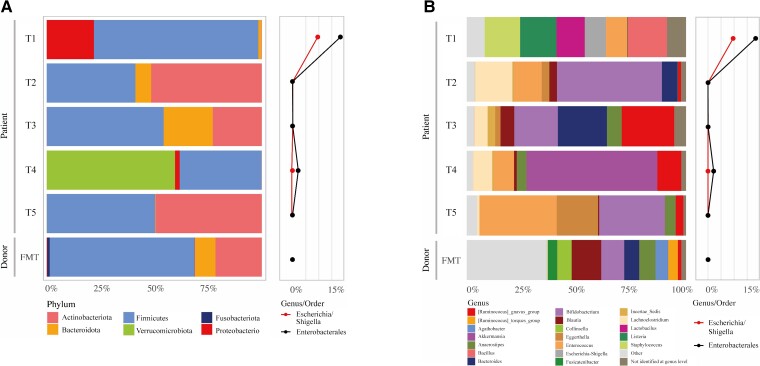
Microbiota analysis was performed by 16S ribosomal RNA (rRNA) gene amplicon sequencing. After DNA extraction from fecal samples, the 16S rRNA gene was amplified by polymerase chain reaction (PCR). The PCR products (amplicons) were then sequenced. Next, the sequences were assigned to bacterial species at the genus level (eg, Klebsiella or Escherichia/Shigella). The protocol for DNA extraction from feces and control samples and the protocol for microbiota analysis are described in the Supplementary Data. Results are shown in Figure 2Aand 2B. Before FMT, the order Enterobacterales was abundant in the recipient’s feces, but was reduced after antibiotic gram-negative decolonization and FMT. Eight months post-FMT, there was a notable increase in the genus Akkermansia. The genus Klebsiella could not be identified in the donor feces or any of the patient samples with this technique.
Figure 2.
Microbiota composition of donor and patient samples at different sampling timepoints, at the phylum level (A) and the genus level (B). Abbreviations: FMT, fecal microbiota transplantation; T1, 4 days pre-FMT; T2, 1 day pre-FMT; T3, 1 month post-FMT; T4, 8 months post-FMT; T5, 17 months post-FMT.
DISCUSSION
A pediatric patient underwent FMT to treat intestinal colonization and multiple recurrent febrile UTIs with MDR K. pneumoniae. The assumed source of the recurrent K. pneumoniae infections was the gut, illustrated by positive fecal cultures after UTI treatment. Reinfection due to infected renal cysts cannot be excluded. However, after FMT, 3 UTIs with E. coli were diagnosed, counterarguing the latter. The E. coli could not be cultured from post-FMT fecal patient samples. However, WGS analysis showed that donor and patient E. coli (from urine) were genetically identical, confirming FMT transmission of E. coli from donor to patient. WGS suggested uropathogenicity of the E. coli by assignment to Clermont phylotype D and the presence of PUFs and additional urovirulence factors [17, 18]. Three months after FMT, the MDR K. pneumoniae recurred in a perineal screening culture: WGS analysis confirmed that this K. pneumoniae was genetically identical to the ones causing UTIs before FMT.
We hypothesize that the K. pneumoniae was temporarily suppressed under the threshold of microbiological detection by the gram-negative gut decolonization (enteral) antibiotics and possibly FMT, but we cannot rule out recolonization from the environment or from a body niche other than the gut. Although FMT was ineffective for resolving recurrent UTIs by MDR K. pneumoniae and decolonization in the long term, several E. coli UTIs after FMT could be successfully treated with oral antibiotics. Our observation confirms that microbiota manipulation has the potential to influence the course of recurrent UTIs. Like Tariq et al, we hypothesize that the course of recurrent UTIs may be changed due to competition and enhanced colonization resistance after the introduction of the donor microbiota [12]. The genus Escherichia/Shigella was detected in low relative abundance (<.02%) by 16S microbiome analysis in patient feces post-FMT and Klebsiella could not be identified at all, possibly due to presence below the level of detection. High abundance of Akkermansia in 1 post-FMT sample might have been the result of broad-spectrum antibiotic use, though fluctuations in absence of antibiotic treatment have also been observed [19]. Antibiotic use post-FMT might also have influenced the restored state of microbiome colonization resistance, allowing for the return of MDRO UTIs.
FMT as a treatment strategy for intestinal eradication of MDROs in pediatric patients has not been previously described in the literature. Here, feces from an adult donor was used. At present, we have no data on whether using feces from a pediatric or adult donor leads to more favorable results. Likewise, the optimal route of FMT administration for this indication is currently unknown.
Previous reports and FDA warnings underline that screening of feces donors via risk assessments and fecal and blood analyses are important to prevent infectious complications [13–15]. The FDA reports focus on MDROs and enteropathogens; however, the decision on which pathogens to screen for is challenging, since translocation of antibiotic-susceptible bacterial gut commensals (including uropathogens) that may be present in both patient and donor may cause infections under specific patient conditions. Not only the presence but also the abundance of certain gut bacteria may be of importance [20]. Furthermore, although many PUFs are described, a clear molecular definition is lacking [18, 21]. Many Enterobacterales contain PUFs, sometimes more abundantly in strains not associated with UTI [18]. Since donor feces screening currently does not include screening for antibiotic-susceptible E. coli and other Enterobacterales, our observation should stimulate more intensive surveillance of post-FMT infections. The exclusion of donor stool based on the mere presence of (antibiotic-susceptible) E. coli is likely not feasible, as we anticipate that many donations would be excluded, with a subsequent impact on donor feces availability and economic feasibility of donor stool programs. The consequences for donor feces screening should be the topic of further studies to enhance FMT safety. Ultimately, standardized preparation of live biotherapeutic products may overcome many of these safety issues.
Supplementary Material
Contributor Information
Karuna E W Vendrik, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands; Netherlands Donor Feces Bank, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands; Centre for Infectious Disease Control, National Institute for Public Health and the Environment, Bilthoven, The Netherlands.
Tim G J de Meij, Department of Pediatrics, Amsterdam University Medical Centers, Emma Children’s Hospital, Amsterdam, The Netherlands.
Arend Bökenkamp, Department of Pediatric Nephrology, Amsterdam Medical Centers, Emma Children’s Hospital, Amsterdam, The Netherlands.
Rogier E Ooijevaar, Netherlands Donor Feces Bank, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands; Department of Gastroenterology and Hepatology, Amsterdam Medical Centers, location Vrije Universiteit University Medical Center, Amsterdam, The Netherlands.
Bas Groenewegen, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands; Netherlands Donor Feces Bank, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands; Center for Microbiome Analyses and Therapeutics, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands.
Antoni P A Hendrickx, Centre for Infectious Disease Control, National Institute for Public Health and the Environment, Bilthoven, The Netherlands.
Elisabeth M Terveer, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands; Netherlands Donor Feces Bank, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands; Center for Microbiome Analyses and Therapeutics, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands.
Ed J Kuijper, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands; Netherlands Donor Feces Bank, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands; Centre for Infectious Disease Control, National Institute for Public Health and the Environment, Bilthoven, The Netherlands; Center for Microbiome Analyses and Therapeutics, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands.
Joffrey van Prehn, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands; Netherlands Donor Feces Bank, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands; Center for Microbiome Analyses and Therapeutics, Department of Medical Microbiology, Leiden University Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. E. M. T., E. J. K., T. G. J. M., A. B., and J. P. conceived the study. K. E. V. and J. P. drafted the manuscript. All authors were involved in data acquisition, analysis, or interpretation, and all authors critically reviewed, revised, and approved the manuscript.
Acknowledgments. The authors thank Rosa van Mansfeld for providing clinical bacterial isolates; Margriet Kraakman for assisting with cgMLST analysis; Eric Berssenbrugge for his contribution to the processing of fecal samples and the storage of Netherlands Donor Feces Bank (NDFB) materials; and Emilie van Lingen for her contribution to the Compassionate Use Program of the NDFB. We also thank the NDFB study group, which consists of Elisabeth M. Terveer, Ed J. Kuijper, Joffrey van Prehn, Vlada Bekker-Chernova, Karuna E. W. Vendrik, and Eline Boeije-Koppenol (Department of Medical Microbiology, Center for Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands); Yvette van Beurden, Rogier Ooijevaar, and Chris J. J. Mulder (Department of Gastroenterology and Hepatology, Amsterdam University Medical Centers, Vrije Universiteit [VU] University Medical Center, Amsterdam, The Netherlands); Merel M. C. Lambregts (Department of Internal Medicine, Leiden University Medical Center, Leiden, The Netherlands); Els van Nood (Department of Internal Medicine, Erasmus Medical Center, Rotterdam, The Netherlands); Abraham Goorhuis (Department of Internal Medicine, Amsterdam University Medical Centers, Amsterdam Medical Center, Amsterdam, The Netherlands); Marcel G. W. Dijkgraaf (Clinical Research Unit, Amsterdam University Medical Centers, Amsterdam, The Netherlands); Christina M. J. E. Vandenbroucke-Grauls (Department of Medical Microbiology and Infection Control, Amsterdam University Medical Centers, VU University Medical Center, Amsterdam, The Netherlands); Hein W. Verspaget (Department of Biobanking and Gastroenterology, Leiden University Medical Center, Leiden, The Netherlands); and Emilie van Lingen and Josbert J. Keller (Department of Gastroenterology, Haaglanden Medical Center, Den Haag, The Netherlands).
Ethics statement. This study was approved by the Medical Ethics Review Committee of Leiden University Medical Center (P15.154).
Data availability. Raw sequence data of the bacterial isolates described in the article can be found in the European Nucleotide Archive (project accession number PRJEB53460).
Financial support. The NDFB has received an unrestricted research grant from Vedanta Biosciences (Cambridge, Massachusetts). There was no specific funding for this study.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Millan B, Park H, Hotte N, et al. Fecal microbial transplants reduce antibiotic-resistant genes in patients with recurrent Clostridium difficile infection. Clin Infect Dis 2016; 62:1479–86. doi: 10.1093/cid/ciw185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuijper EJ, Vendrik KEW, Vehreschild M. Manipulation of the microbiota to eradicate multidrug-resistant Enterobacteriaceae from the human intestinal tract. Clin Microbiol Infect 2019; 25:786–9. doi: 10.1016/j.cmi.2019.03.025 [DOI] [PubMed] [Google Scholar]
- 3. Huttner BD, de Lastours V, Wassenberg M, et al. A 5-day course of oral antibiotics followed by faecal transplantation to eradicate carriage of multidrug-resistant Enterobacteriaceae: a randomized clinical trial. Clin Microbiol Infect 2019; 25:830–8. doi: 10.1016/j.cmi.2018.12.009 [DOI] [PubMed] [Google Scholar]
- 4. Worby CJ, Schreiber HLT, Straub TJ, et al. Longitudinal multi-omics analyses link gut microbiome dysbiosis with recurrent urinary tract infections in women. Nat Microbiol 2022; 7:630–9. doi: 10.1038/s41564-022-01107-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stalenhoef JE, Terveer EM, Knetsch CW, et al. Fecal microbiota transfer for multidrug-resistant gram-negatives: a clinical success combined with microbiological failure. Open Forum Infect Dis 2017; 4:ofx047. doi: 10.1093/ofid/ofx047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biehl LM, Cruz Aguilar R, Farowski F, et al. Fecal microbiota transplantation in a kidney transplant recipient with recurrent urinary tract infection. Infection 2018; 46:871–4. doi: 10.1007/s15010-018-1190-9 [DOI] [PubMed] [Google Scholar]
- 7. Wang T, Kraft CS, Woodworth MH, Dhere T, Eaton ME. Fecal microbiota transplant for refractory Clostridium difficile infection interrupts 25-year history of recurrent urinary tract infections. Open Forum Infect Dis 2018; 5:ofy016. doi: 10.1093/ofid/ofy016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hocquart M, Pham T, Kuete E, Tomei E, Lagier JC, Raoult D. Successful fecal microbiota transplantation in a patient suffering from irritable bowel syndrome and recurrent urinary tract infections. Open Forum Infect Dis 2019; 6:ofz398. doi: 10.1093/ofid/ofz398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grosen AK, Povlsen JV, Lemming LE, Jørgensen SMD, Dahlerup JF, Hvas CL. Faecal microbiota transplantation eradicated extended-spectrum beta-lactamase–producing Klebsiella pneumoniae from a renal transplant recipient with recurrent urinary tract infections. Case Rep Nephrol Dial 2019; 9:102–7. doi: 10.1159/000502336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aira A, Rubio E, Vergara Gómez A, et al. rUTI resolution after FMT for Clostridioides difficile infection: a case report. Infect Dis Ther 2020; 10:1065–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jeney SES, Lane F, Oliver A, Whiteson K, Dutta S. Fecal microbiota transplantation for the treatment of refractory recurrent urinary tract infection. Obstet Gynecol 2020; 136:771–3. doi: 10.1097/AOG.0000000000004052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tariq R, Pardi DS, Tosh PK, Walker RC, Razonable RR, Khanna S. Fecal microbiota transplantation for recurrent Clostridium difficile infection reduces recurrent urinary tract infection frequency. Clin Infect Dis 2017; 65:1745–7. doi: 10.1093/cid/cix618 [DOI] [PubMed] [Google Scholar]
- 13. DeFilipp Z, Bloom PP, Torres Soto M, et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med 2019; 381:2043–50. doi: 10.1056/NEJMoa1910437 [DOI] [PubMed] [Google Scholar]
- 14. Zellmer C, Sater MRA, Huntley MH, Osman M, Olesen SW, Ramakrishna B. Shiga toxin-producing Escherichia coli transmission via fecal microbiota transplant. Clin Infect Dis 2021; 72:E876–80. doi: 10.1093/cid/ciaa1486 [DOI] [PubMed] [Google Scholar]
- 15. Vendrik KEW, Terveer EM, Kuijper EJ, et al. Periodic screening of donor faeces with a quarantine period to prevent transmission of multidrug-resistant organisms during faecal microbiota transplantation: a retrospective cohort study. Lancet Infect Dis 2021; 21:711–21. doi: 10.1016/S1473-3099(20)30473-4 [DOI] [PubMed] [Google Scholar]
- 16. Hendrickx APA, Landman F, de Haan A, et al. bla OXA-48-like genome architecture among carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the Netherlands. Microb Genom 2021; 7:000512. doi: 10.1099/mgen.0.000512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microb 2000; 66:4555–8. doi: 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schreiber HLT, Conover MS, Chou WC, et al. Bacterial virulence phenotypes of Escherichia coli and host susceptibility determine risk for urinary tract infections. Sci Transl Med 2017; 9:eaaf1283. doi: 10.1126/scitranslmed.aaf1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stewardson AJ, Gaia N, Francois P, et al. Collateral damage from oral ciprofloxacin versus nitrofurantoin in outpatients with urinary tract infections: a culture-free analysis of gut microbiota. Clin Microbiol Infect 2015; 21:344.e1–11. doi: 10.1016/j.cmi.2014.11.016 [DOI] [PubMed] [Google Scholar]
- 20. Magruder M, Sholi AN, Gong C, et al. Gut uropathogen abundance is a risk factor for development of bacteriuria and urinary tract infection. Nat Commun 2019; 10:5521. doi: 10.1038/s41467-019-13467-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spurbeck RR, Dinh PC Jr, Walk ST, et al. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect Immun 2012; 80:4115–22. doi: 10.1128/IAI.00752-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.