Figure 5.
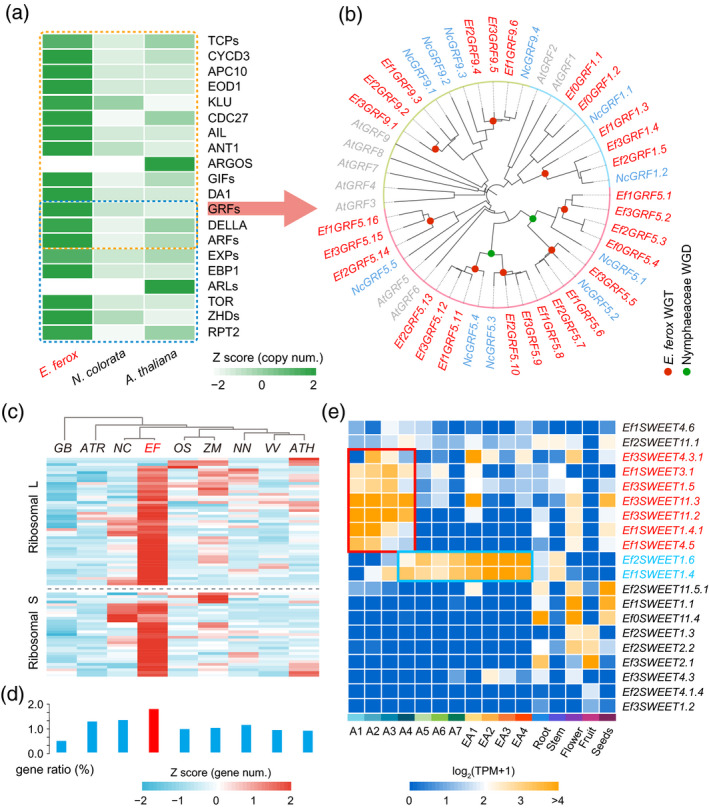
Gene families associated with the specific development of leaves in Euryale ferox.
(a) The expansion of gene families related to cell expansion and division in E. ferox, compared with that in Nymphaea colorata and Arabidopsis.
(b) Phylogenetic tree of GRF genes in E. ferox, N. colorata and Arabidopsis. Red denotes E. ferox genes, green denotes N. colorata genes, and gray denotes Arabidopsis genes.
(c) Comparisons of the number of ribosomal protein (RP) gene families in Ginkgo biloba (Gb), Amborella trichopoda (Atr), N. colorata (Nc), E. ferox (Ef), Oryza sativa (Os), Zea mays (Zm), Nelumbo nucifera (Nn), Vitis vinifera (Vv) and Arabidopsis (Ath).
(d) Ratio of the number of RP genes to the number of total genes in the corresponding genome.
(e) Expression atlases of SWEET genes in the leaves of different developmental stages and other organs of E. ferox. [Colour figure can be viewed at wileyonlinelibrary.com]
