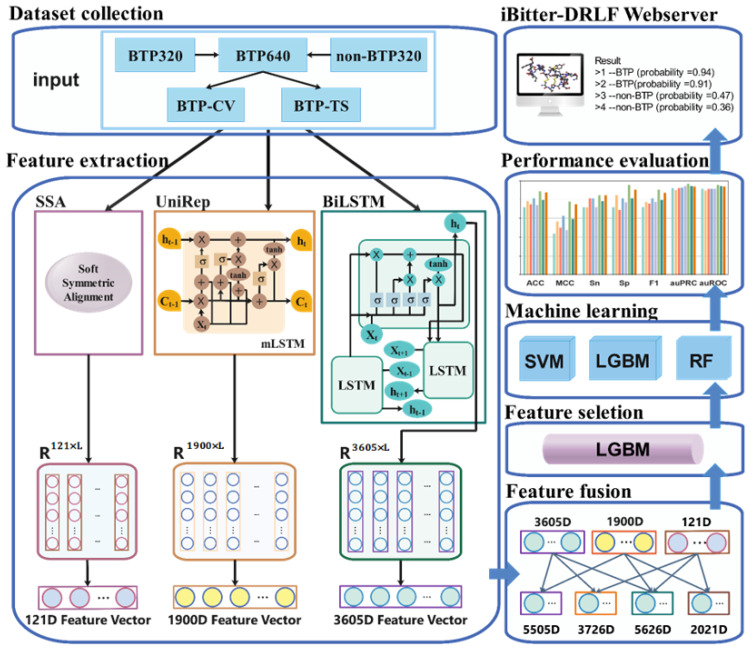
Figure 1.
Overview of model development. The pre-trained SSA sequence embedding model, UniRep sequence embedding model, and BiLSTM sequence embedding model were used to embed peptide sequences into eigenvectors. Peptide sequences were converted into 121-dimensional (D) SSA eigenvectors, 1900-dimensional UniRep eigenvectors, and 3605-dimensional (D) BiLSTM eigenvectors. Features were combined and fused to derive the following fusion features: SSA-UniRep (2021D), SSA-BiLSTM (3726D), UniRep-BiLSTM (5505D), and SSA-UniRep-BiLSTM (5626D). These fusion features were used as inputs to the SVM, RF, and LGBM predictor algorithms. The model was optimized by feature selection using the LGBM method. Selected feature sets were subjected to another round of analysis using three algorithms and various hyperparameters. Through 10-fold cross-validation and comparison of independent tests results, the optimized final model was developed. Here, the example like SSA-BiLSTM means two kind of features are combined.