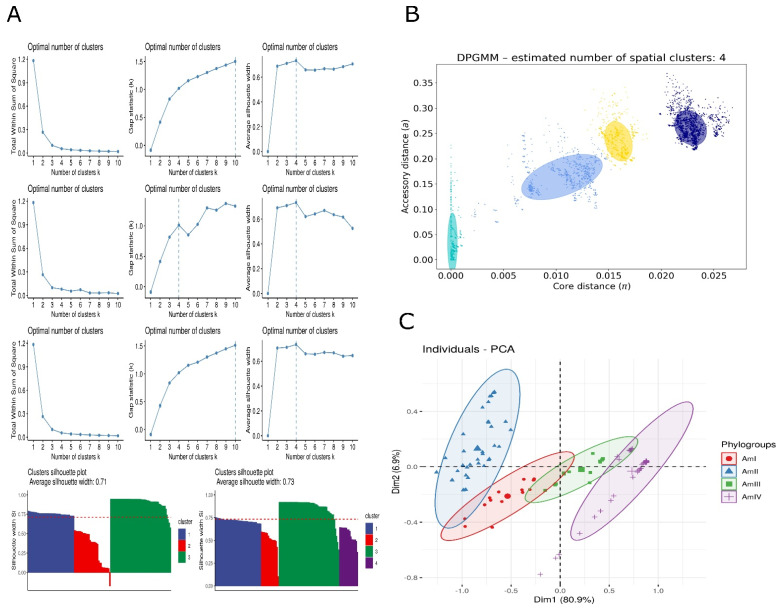
Figure 4.
Clustering of strains into phylogroups. (A) (I–III) Partition around medoids clustering prediction using (I) within sum of square, (II) gap distances, and (III) silhouette distances. (IV–VI) Kmeans clustering prediction using (IV) within sum of square, (V) gap distances, and (VI) silhouette distances. (VII–IX) Hierarchical clustering prediction using (VII) within sum of square, (VIII) gap distances (IX), and silhouette distances. (X) Silhouette plot for three clusters. (XI) Silhouette plot for four clusters. (B) Dirichlet Process Gaussian Mixture Model-based clustering of strains into phylogroups using core distances and accessory distances. (C) Principal component analysis of gene presence-absence profile.