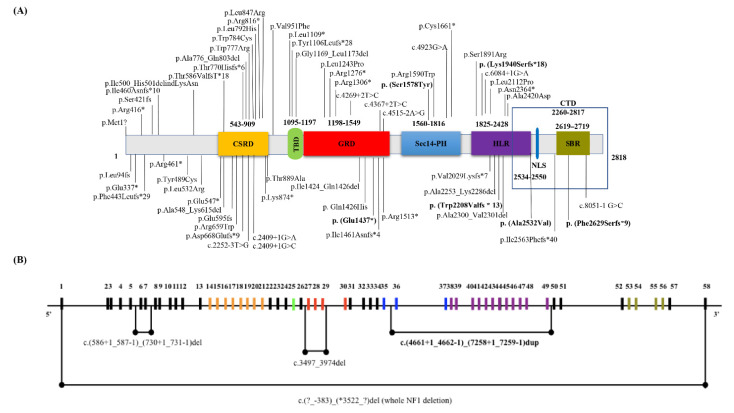
Figure 1.
(A) Distribution of the identified mutations in the NF1 gene. Details of the 66 NF1 genetic mutations identified in our NF1 Italian cohort. The position of genetic variations detected in the NF1 from each NF1 patient is shown and their distribution in the NF1 domains is reported. Vertical lines show variant position. NF1 novel variants are shown in bold. Neurofibromin domains: CSRD, cysteine/serine-rich domain (543–909 residues); GRD, GAP related domain (1198–1549 residues); TBD, tubulin-binding domain (1095–1197 residues); HLR, HEAT-like repeat regions (1825–2428 residues); NLS, nuclear localization signal (2534–2550 residues); Sec14-PH, Sec14-homologous domain (1560–1816 residues) and Pleckstrin Homology domain (1716–1816 residues); CTD, C-terminal domain (2260–2817 residues); SBR, Syndecan-Binding Region (2619–2719 residues). (B) Map of the NF1 region indicating the large duplications and deletions sequences identified in our NF1 cohort. The black horizontal line represents the entire NF1 gene, while the vertical lines indicate the NF1 exons. Different colors were used for NF1 exons according to the relative functional domains as shown in (A).