Figure 4: Loss of Mef2c disproportionately affects the gene regulatory landscape of PV cells.
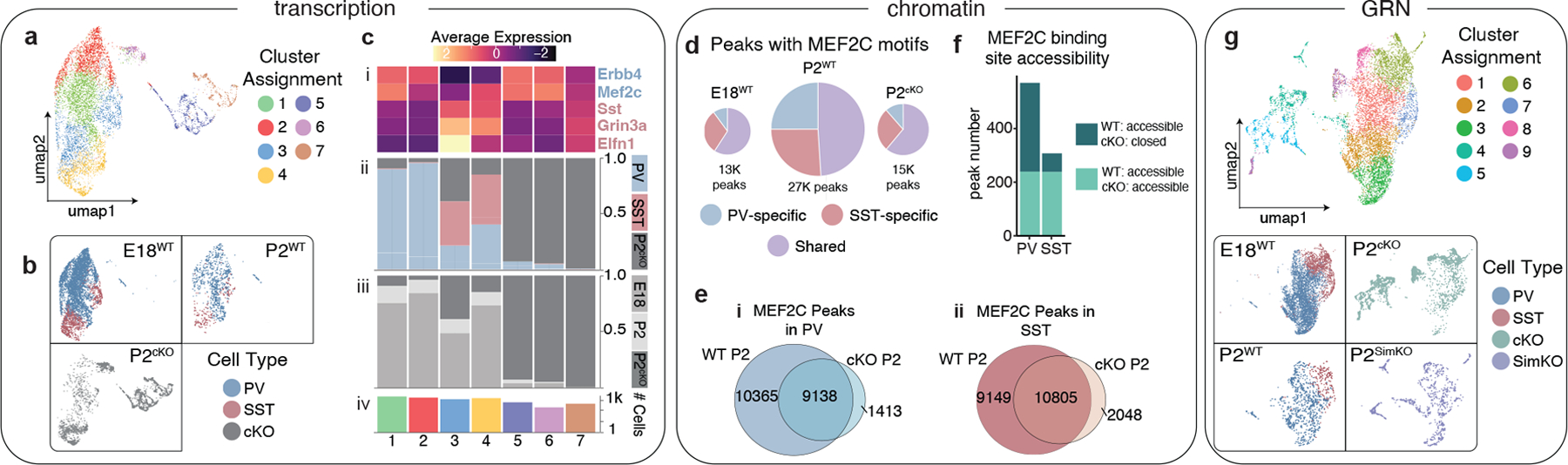
a, UMAP of integrated E18, P2 and P2 Mef2c cKO scRNA-seq data.
b, UMAP in (a) segregated according to timepoint and color-coded by cell type.
c, Cluster composition delineated by (i) marker gene expression, (ii) cell type, (iii) timepoint, (iv) cell number.
d, Proportion of scATAC-seq peaks with MEF2C motifs in E18 WT, P2 WT, and P2 Mef2c cKO datasets. Charts are scaled to reflect the total number of peaks.
e, Venn diagram showing all peaks with MEF2C motifs in WT P2 and cKO P2 (i) in PV cells, (ii) SST cells.
f, Number of MEF2C binding sites (identified in the MEF2C CUT&RUN experiment, see Fig. 2) that were either accessible in both the P2 WT and Mef2c cKO scATAC-seq dataset (light green) or that were accessible in the P2 WT cells but inaccessible in the cKO (dark green).
g, UMAP of simulated Mef2c knockout cells (P2SimKO) and cells from the true cKO RNA-seq dataset (P2cKO), P2 WT RNA-seq dataset (P2WT), and E18 WT RNA-seq dataset (E18).
