Extended Data Figure 1. Quality control of embryonic scRNA-seq samples and mitotic versus postmitotic discrimination.
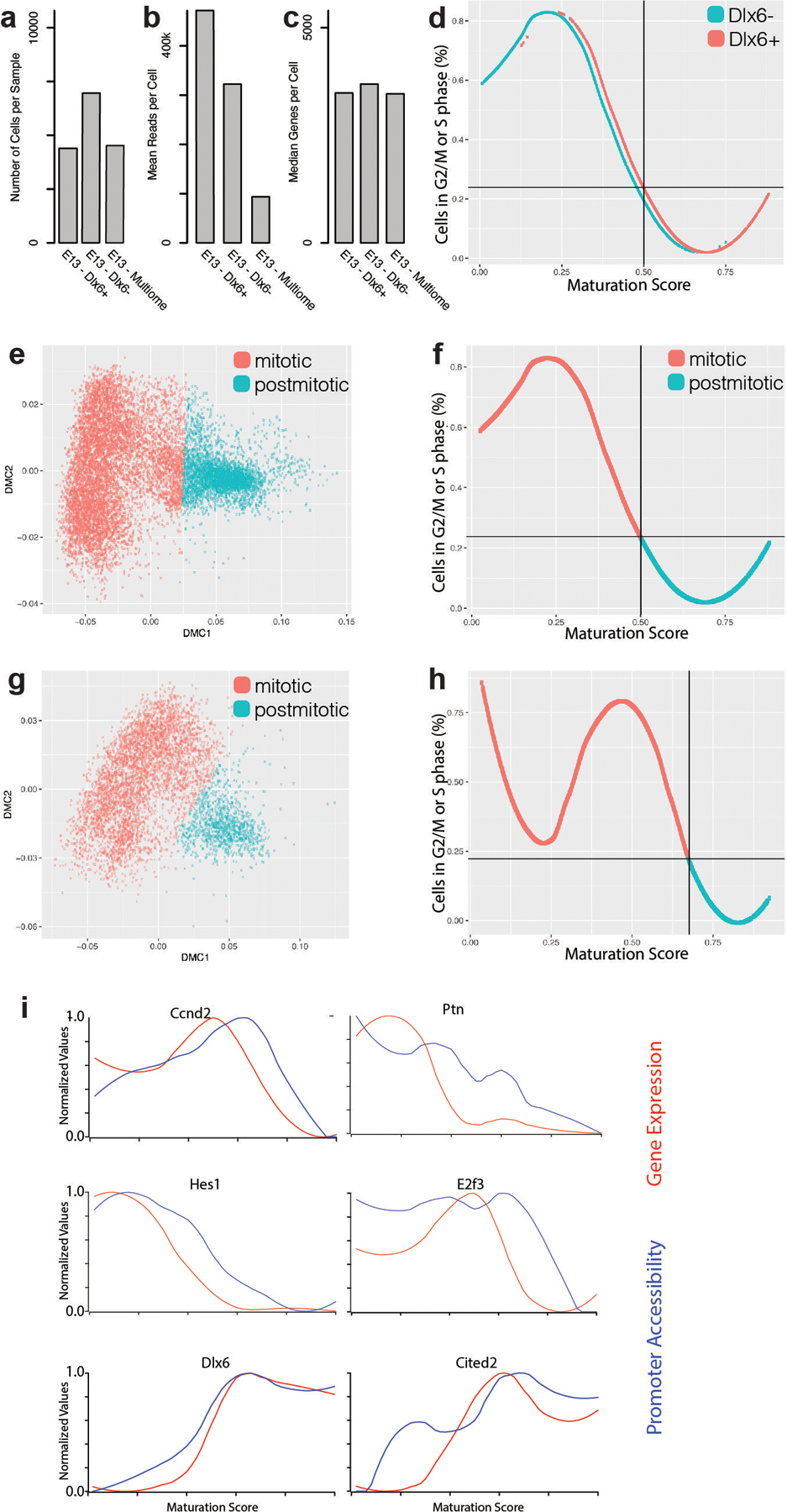
a, Number of cells in Dlx6a− and Dlx6a+ scRNA-seq datasets collected from E13 MGE in Dlx6a-Cre;Ai9 mice and multiome dataset in E13 MGE wild type mice.
b, Mean reads per cell in Dlx6a−, Dlx6a+ and multiome scRNA-seq datasets.
c, Median genes detected per cell in Dlx6a−, Dlx6a+ and multiome scRNA-seq datasets.
d, Fraction of cells scored to be in G2/M or S phase of the cycle cycle at each maturation score. Blue line indicates cells from the Dlx6- dataset, red line indicates cells from the Dlx6+ dataset.
e, Diffusion map of E13 Dlx6a− and Dlx6a+ MGE scRNA-seq data color-coded by assignment to a mitotic (red) or postmitotic (blue) state.
f, Percentage of cycling cells as a function of the position along the maturation trajectory color-coded by assignment to a mitotic (red) or postmitotic (blue) state in E13 Dlx6a− and Dlx6a+ MGE scRNA-seq datasets.
g, Diffusion map of E13 multiome MGE scRNA-seq data color-coded by assignment to a mitotic (red) or postmitotic (blue) state.
h, Percentage of cycling cells as a function of the position along the maturation trajectory color-coded by assignment to a mitotic (red) or postmitotic (blue) state in E13 multiome MGE scRNA-seq dataset.
i, Line plots indicating the promoter accessibility (blue) and gene expression (red) for six developmentally-regulated genes.
