Extended Data Figure 2. Developmental characterization of embryonic E13 MGE cells surveyed using scRNA-seq, scATAC-seq, and multiomic methods.
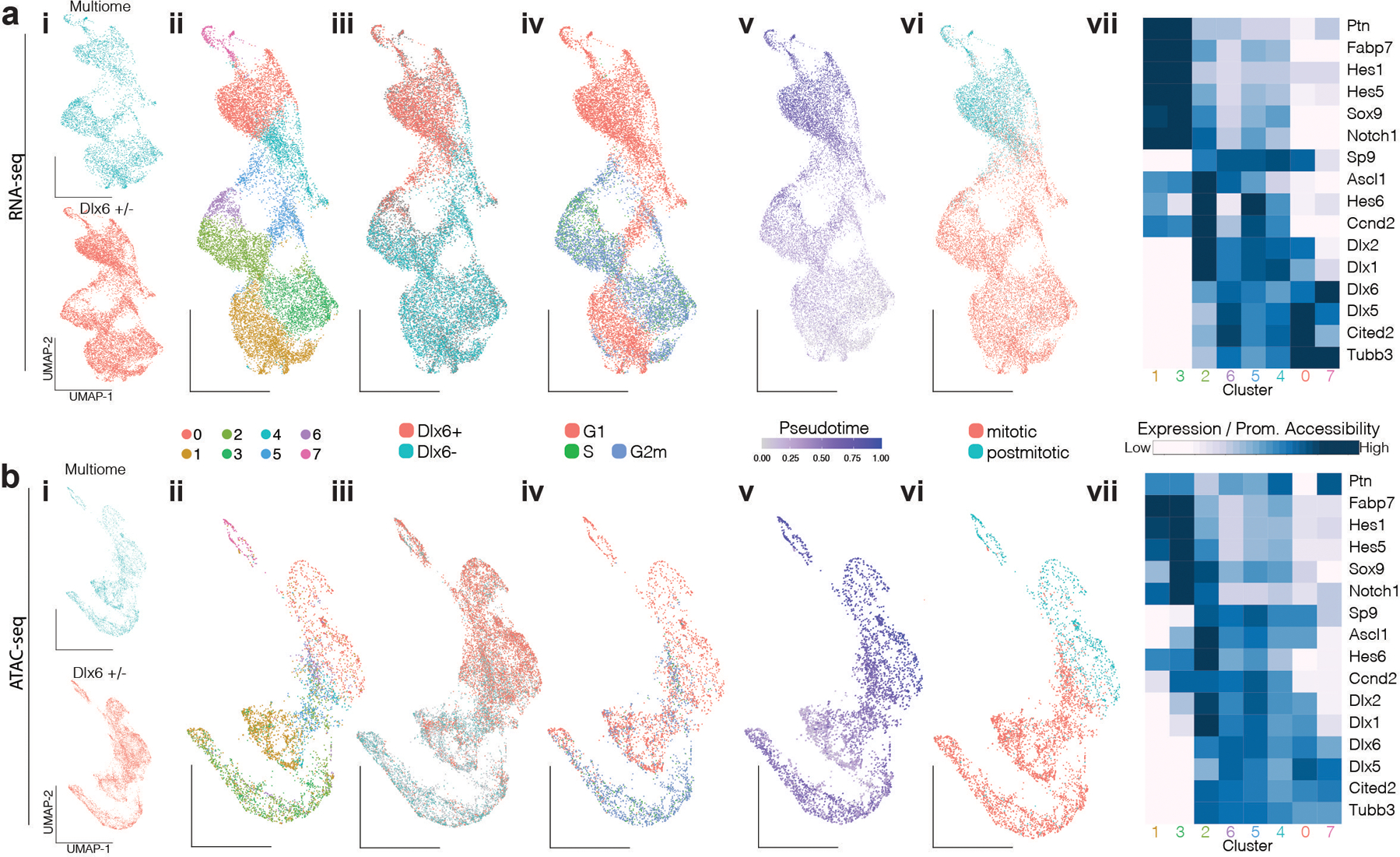
a, Analysis of Dlx6a−, Dlx6a+ and multiome scRNA-seq datasets collected from E13 MGE in Dlx6a-Cre;Ai9 mice and multiome dataset in E13 MGE wild type mice.
b, Analysis of Dlx6a−, Dlx6a+ and multiome scATAC-seq datasets collected from E13 MGE in Dlx6a-Cre;Ai9 mice and multiome dataset in E13 MGE wild type mice.
i, Dlx6+/− FAC-sorted and multiome cells.
ii, Unbiased cluster annotation.
iii, Dlx6+ and Dlx6- annotation.
iv, Cell cycling phase annotation.
v, Pseudotime annotation.
vi, Mitotic and postmitotic cell annotation.
In ii-vi, Annotations are performed on scRNA-seq datasets and transferred to scATAC-seq datasets through the multiome dataset. scRNA- and scATAC-seq low dimensional representation reflects UMAP embedding.
vii, Average gene expression and promoter accessibility for unbiased clusters.
