Extended Data Figure 4. Chromatin accessibility precedes gene expression in branch 1- specific genes.
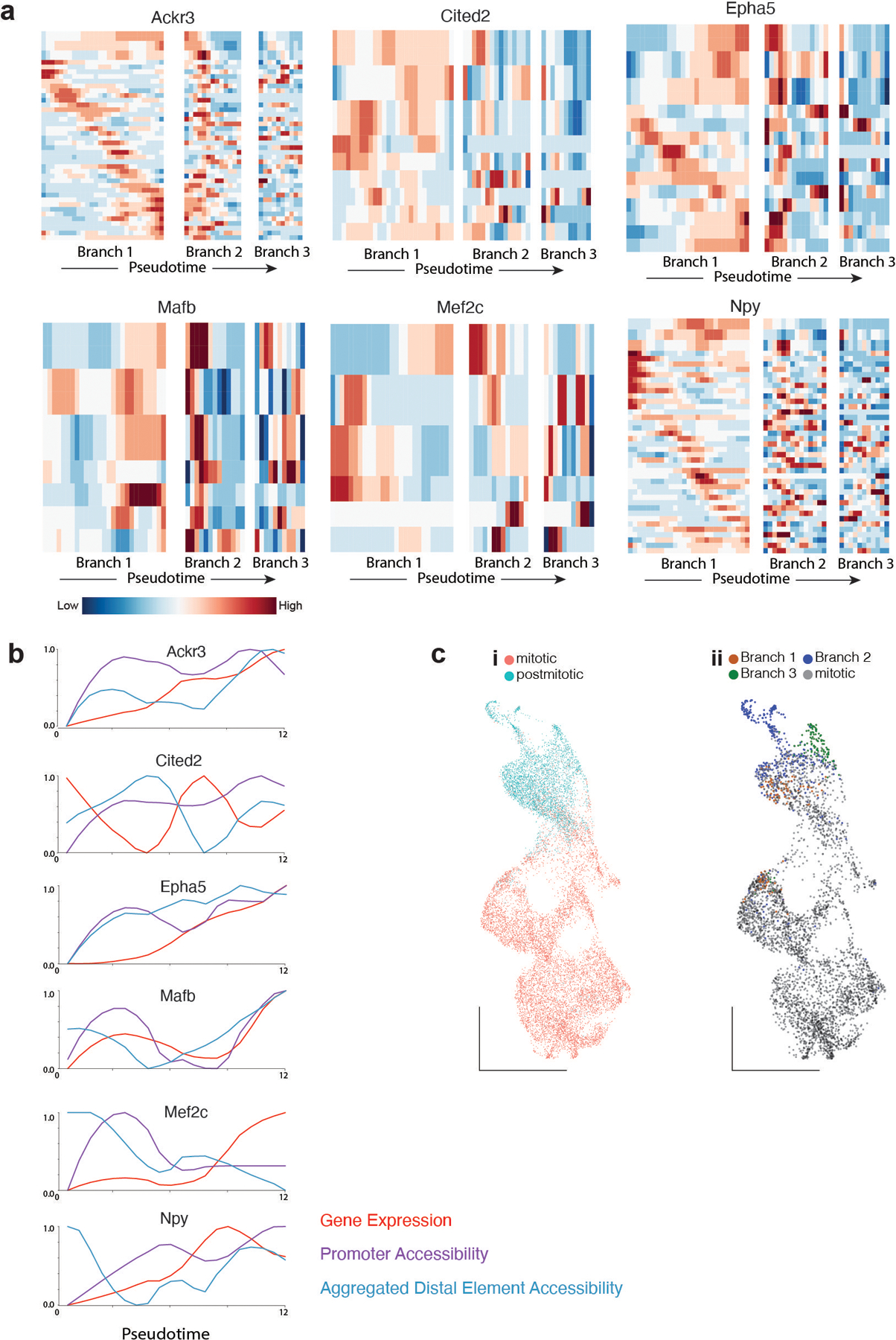
a, Heatmap depicting gene expression, Promoter and distal elements accessibility throughout branch 1 pseudotime for loci +/− 500kb around six branch 1 - specific genes. Distal elements are selected based on the relevance for classifying branch 1 cells. For each gene, row 1 shows gene expression, row 2 promoter accessibility, row 3 aggregated accessibility and the remaining rows are distal branch 1 classifying elements. Each trace has been smoothed using the lowess function in R.
b, Gene expression, Promoter and Aggregated accessibility throughout branch 1 pseudotime for 10 branch 1 - specific genes.
c, Only postmitotic cells are classified as branch neurons by supervised classification methods.
c-i) Classification of E13 cells into mitotic or postmitotic cells based on cell cycle RNA score.
c-ii) Classification of E13 cells into mitotic or branch 1,2,3 lineages based on chromatin accessibility.
