Extended Data Figure 5: Analysis of MGE-derived cortical interneuron scRNA-seq datasets from E18 through P28.
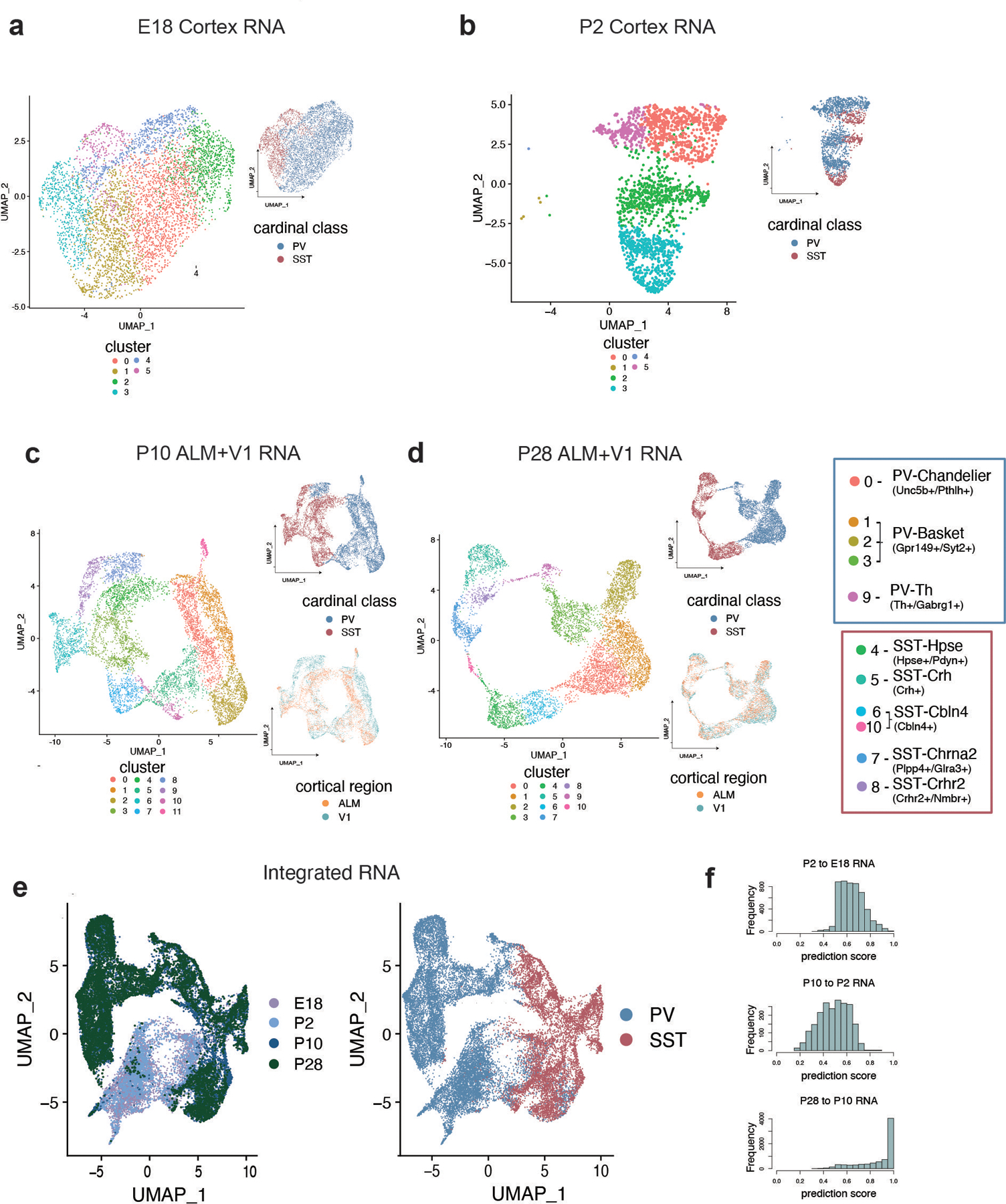
a-d, UMAP showing individual scRNA-seq datasets from Dlx6a+ labeled cortical neurons, subsetted for PV+ and SST+ interneurons, collected at E18 (a), P2 (b), P10 (c), and P28 (d). Left UMAP in each panel is color-coded by cluster identity (individually determined for each dataset). Right UMAP in each panel is color-coded by cardinal class. Bottom right UMAP in (c) and (d) is color-coded by cortical region of origin (anterior lateral motor cortex - ALM, primary visual cortex - V1). In (d), the right-most boxes show each cluster linked to its subtype identity, determined by expression of marker genes.
e, Integration of E18, P2, P10, and P28 scRNA-seq datasets using Seurat CCA. Left UMAP is color-coded by timepoint, right is color-coded by cardinal class.
f, Prediction score indicating the confidence of label transfer between each timepoint. Cluster labels were transferred from P2 to E18 (top), from P10 to P2 (middle) and from P28 and P10 (bottom). Label transfer prediction scores were lowest between P10 and P2 timepoints.
