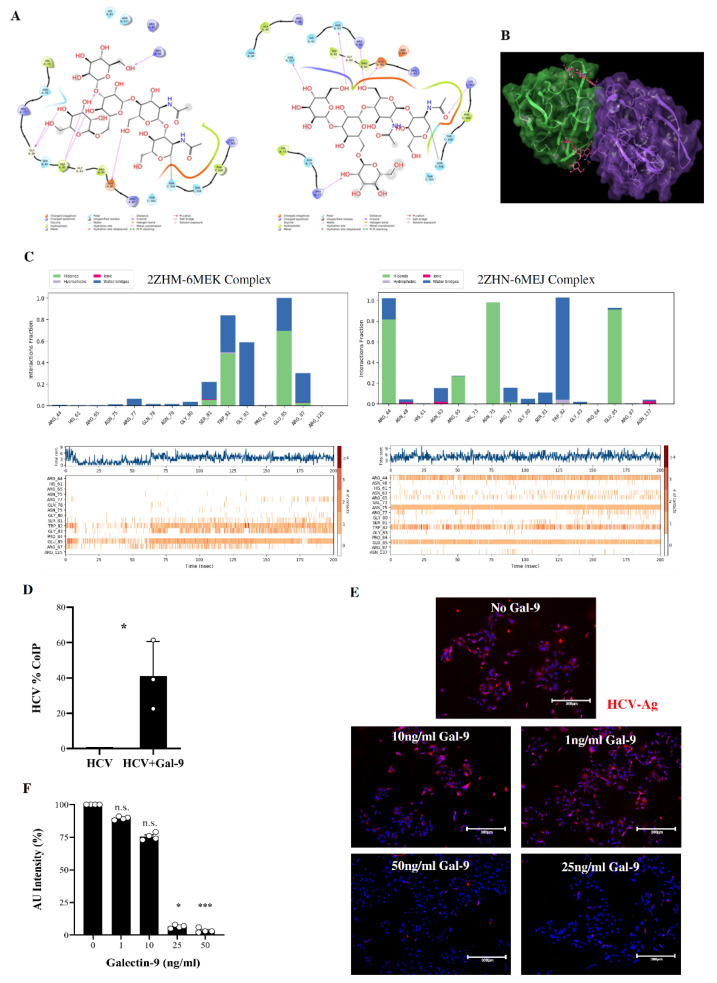
Figure 6.
In silico and functional characterization of neutralizing interactions between HCV and galectin-9. (A) On the left, ligand interaction diagram of HCV E2 carbohydrate from PDB 6MEJ and galectin-9 CRDs from PDB 2ZHM; on the right, ligand interaction diagram of HCV E2 carbohydrate from PDB 6MEK and galectin-9 CRDs from PDB 2ZHN. (B) Galectin-9 CRDs–HCV E2 complex retrieved from a selected docking pose. The green chain is the galectin-9 CRDs from PDB 2ZHN, the purple chain represents HCV E2 from PDB 6MEJ, and the pink molecules are the sugar moieties belonging to E2. (C) On the top, bar plots of the interactions established between galectin-9 CRDs and HCV E2 during the MD trajectories; on the bottom, frequency occurrence of the interactions between the protein partners in the MD timeframe. (D) Bar graph showing the percentage of HCV bound to galectin-9, as assessed with the co-immunoprecipitation (CoIP) assay. HCV without galectin-9 was used as a negative control. Each dot represents a technical replicate. Statistical analyses were performed using Student’s t-test (p-value * ≤ 0.05). (E) Representative immunofluorescence staining of HCV-Ag (red color) in Huh7.5 cells infected with recombinant HCV preincubated with decreasing doses (50–0 ng/mL) of galectin-9. DAPI (blue color) was used to counterstain the nuclei. Magnification of 200×. (F) Intensity of fluorescent HCV-Ag signal in cells as in A, expressed as percentage of arbitrary units (AU), as quantified with ImageJ software. Each dot represents an independent experiment (n = 4). Mean values ± SEM are shown. Statistical analyses were performed using the Kruskal–Wallis test followed by Dunn’s multiple comparison test. * p < 0.05, *** p < 0.001; n.s., non-significant.