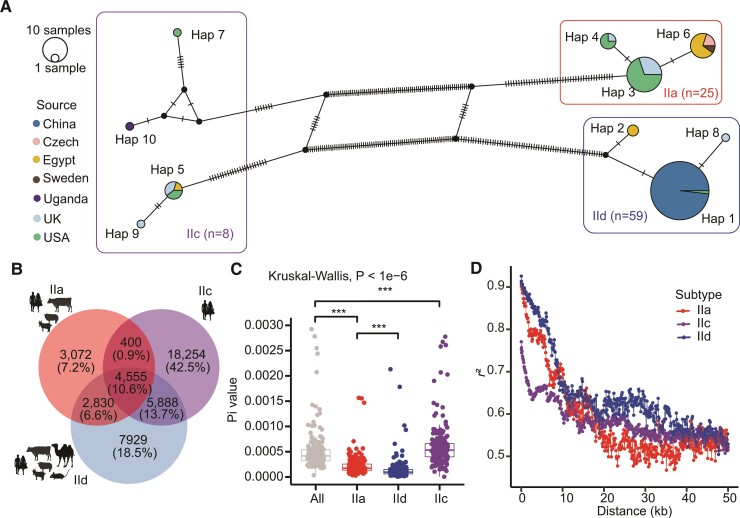
Fig. 1.
Genomic differences among three major gp60 subtype families of Cryptosporidium parvum. (A) Haplotype network of gp60 sequences extracted from 101 genomes used in the study, including those of IIa, IIc, and IId subtype families from China, Egypt, the Czech Republic, Sweden, Uganda, the United Kingdom, and the United States. Each circle indicates a unique haplotype of C. parvum with the color corresponding to the geographic origin. The size of the circle is proportional to the number of samples with the haplotype. (B) Distribution of shared and subtype family-specific SNPs among three gp60 subtype families with different host ranges, including IIa (mainly found in humans, cattle, sheep, and goats), IIc (mainly found in humans), and IId (found in humans, farm animals, and small rodents). (C) Nucleotide divergence (Pi) within each gp60 subtype family. The Pi values were determined with a 50 kb sliding window along the genome for isolates of four groups, including IIa, IIc, IId, and all samples (***P < 0.001). (D) Decay of genome-wide LD by subtype family. LD between SNPs within a 50 kb distance was calculated for IIa, IIc, and IId subtype families. An equal number of samples were randomly selected in each subtype family for LD calculation to reduce the potential influence of the sample size. Resampling was performed 50 times and the mean values obtained were used in plotting.