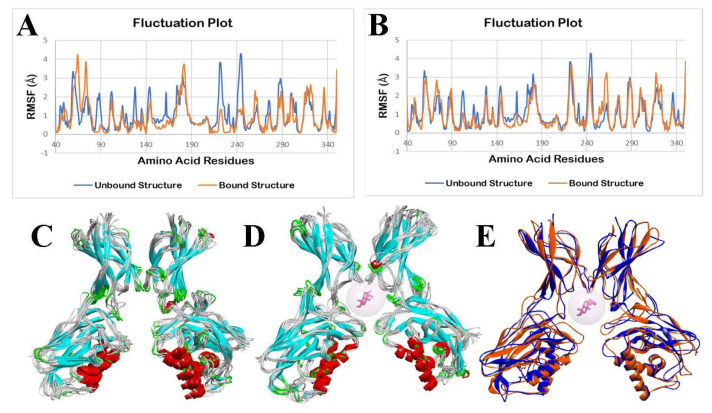
Figure 19.
Molecular dynamics simulation of unbound and 14−deoxy−14,15−dehydroandrographolide bound NF−κB p50 structures: (A) fluctuation plot of the number of amino acid residues of unbound (blue) and bound structure (orange) of NF−κB p50 chain A, (B) fluctuation plot of the number of amino acid residues of unbound (blue) and bound structure (orange) of NF−κB p50 chain B, (C) superimposition of unbound NF−κB p50 structures from 0 ns to 10 ns, (D) superimposition of 14−deoxy14,15−dehydroandrographolide bound NF−κB p50 structures from 0 ns to 10 ns, and (E) superimposition of unbound NF-κB p50 at 0 ns and 14−deoxy−14,15−dehydroandrographolide bound NF−κB p50 at 10 ns.