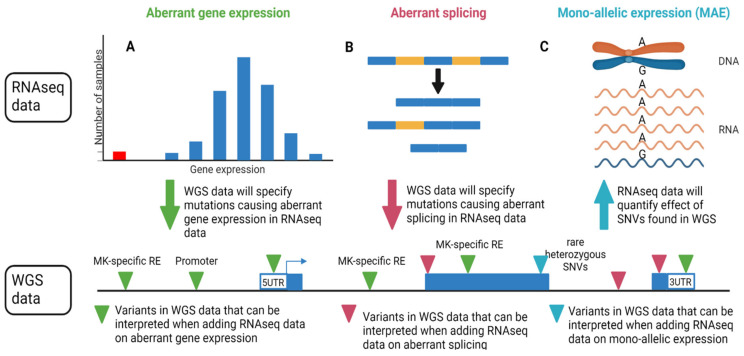
Figure 2.
Schematic of information gained by combining RNA-seq data and Whole Genome Sequencing (WGS) data. (A) RNA-seq gene expression outliers offer insight into the transcriptomic effect of coding and non-coding patient mutations in WGS data. (B) The probability of a change in DNA sequence causing alternative splicing can be predicted by deep learning. Predictions made for patient mutations seen in WGS data can be verified through splicing outlier detection in RNA-seq data. (C) The effect of heterogeneous patient mutations from WGS data is also dependent on monoallelic expression, in which one of the two alleles is (partially) unused. This can be quantified using RNA-seq data.