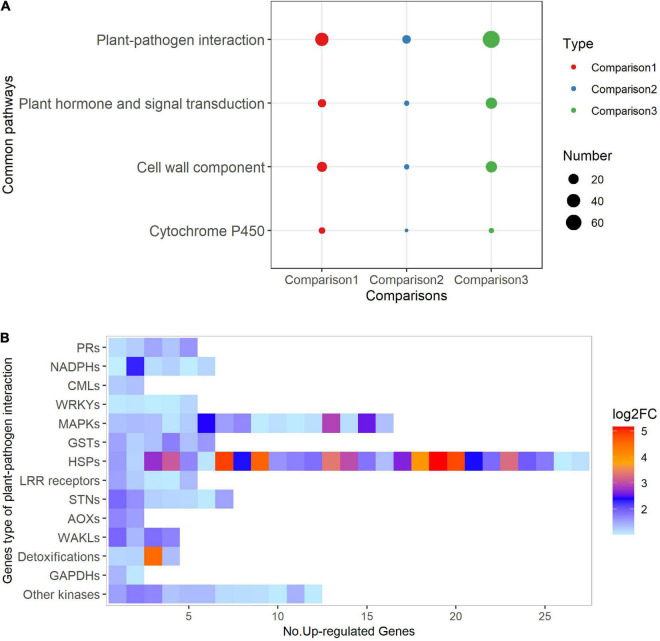
FIGURE 4.
Functional pathways analysis of up-regulated genes. (A) Common pathways identified in up-regulated genes in all three transcriptome comparisons. Comparison1, 48 DAS_ J2416K vs. J2416; Comparison2, 62 DAS_ J2416K vs. J2416; Comparison3, J2416K_ 62 DAS vs. 48 DAS. (B) Heatmap of 14 types of genes associated with the plant–pathogen interaction pathway. PRs, pathogenesis-related genes; NADHs, NADPH oxidases; CMLs, CaM-like proteins; WRKYs, WRKY transcription factors; MAPKs, mitogen-activated protein kinases; GSTs, glutathione transferases; HSPs, heat shock proteins; STNs, serine/threonine protein kinases; AOXs, alternative oxidases; WAKLs, wall-associated receptor kinases; GAPDHs, glyceraldehyde-3-phosphate dehydrogenases.