Abstract
The objective of this study is to report the complete-genome sequence of a field African swine fever (ASF) virus (ASFV), namely ASF/VN/CanTho-OM/2021, which caused a fatal outbreak in domestic pigs in the Mekong Delta. Complete-genome sequencing detected an 18 bp nucleotide deletion in the EP402R gene (encoding for serotype-specific proteins CD2v) of ASF/VN/CanTho-OM/2021, which was determined to belong to genotype 2 and serotype 8. This mutation pattern was confirmed as unique in GenBank; thus, ASF/VN/CanTho-OM/2021 can be considered a novel variant, with a potential change of sero-characteristics within genotype 2. An additional unique mutation of 78 bp nucleotide insertion was also observed in the B475L gene. Additionally, four copies of tandem repeat sequences were found in the intergenic region (IGR) located between I73R and I329L, previously assigned as the IGR III variant. This study is the first to report the complete genome of ASFV in the Mekong Delta, and it highlights the necessity of strengthening molecular surveillance to provide further knowledge on the evolution and incursion of ASFV in the Mekong Delta and Vietnam.
Keywords: African swine fever, complete genome, Mekong Delta, variant, Vietnam
1. Introduction
African swine fever (ASF) is a highly infectious disease in pigs caused by the African swine fever virus (ASFV), which might result in a high mortality rate approaching 100% [1,2]. ASFV is the sole member of the family Asfarviridae, a DNA arbovirus. The viral genome of ASFV has 170 to 195 kbp, encoding at least 160 open reading frames [3,4]. Currently, 24 genotypes and 8 serotypes have been reported worldwide based on the B646L gene encoding the major capsid protein p72 and the EP402R gene encoding the viral hemagglutinin CD2-like protein (CD2v) [5].
ASF was first reported in Kenya in 1921 [6]. Since the 1950s, ASFV has spread throughout Europe in two separate epidemic waves. The first wave in 1957 was through Spain and Portugal to other countries in Western Europe, and it was eradicated in many countries by the mid-1990s [1,7]. The second wave of the disease started in 2007, when ASF was detected in Georgia, and subsequently in neighboring countries in Eastern Europe; since then, ASF has become endemic in Russia and several European countries [8]. Since its first introduction to China in August 2018, ASF has spread rapidly to several other Asian countries [9]. Specifically, in Vietnam, the disease was first reported in February 2019 in the Northern provinces and became endemic afterward, causing serial outbreaks in domestic pigs across the country. Several previous studies have been conducted to determine the genotypes and serotypes of ASFV circulating in Vietnam and have confirmed that all detected ASFVs belong to genotype 2 and serotype 8 [10,11,12]. Currently, at least three variants of genotype 2 based on the variation of tandem repeat sequences (TRS) in the intergenic region (IGR) located between I73R and I329L genes have been identified in North Vietnam [13,14]. Moreover, other mutations might exist in the genome of circulating ASFVs, which might alter the virulence and sero-characteristics of field viruses; nevertheless, information on the complete genome of field ASFVs in Vietnam remains limited. Therefore, in this study, a causative ASFV of an outbreak in domestic pigs in the Mekong Delta in 2021 was subjected to next-generation sequencing (NGS), and this study revealed a novel variant based on EP402R (encoding for serotype-specific proteins CD2v) and B475L of the field ASFV.
2. Results
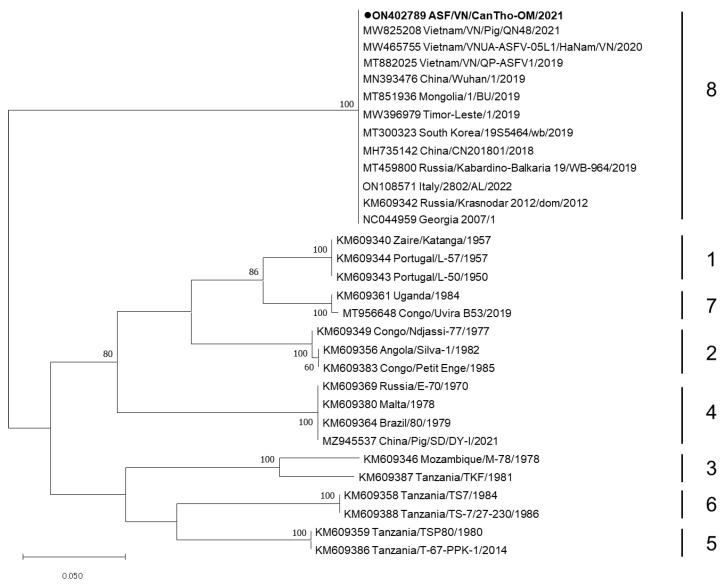
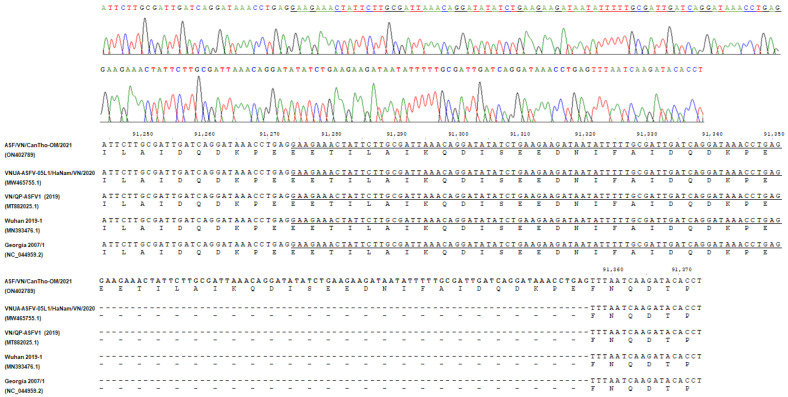
The whole-genome sequence (WGS) of ASFV detected in this study, namely ASF/VN/CanTho-OM/2021, was deposited to GenBank and assigned the accession number ON402789. Based on the sequence alignment of the B646L gene (p72), ASF/VN/CanTho-OM/2021 showed 100% nucleotide identity with ASFVs previously detected in Vietnam, China and Georgia 2007/1, which all belong to genotype 2 (data not shown). Similarly, a phylogenetic analysis of the EP402R gene (CD2v) confirmed that ASF/VN/CanTho-OM/2021 belongs to serotype 8 (Figure 1). Interestingly, ASF/VN/CanTho-OM/2021 had an 18 bp nucleotide deletion of “CTACTACCCAATATCCCG” at positions 75,614 to 75,631 (based on the numbering scheme of NC_044959.2, Georgia 2007/1, Figure 2). The new sequence pattern of EP402R was confirmed as unique because there was no homology to other sequences previously deposited in GenBank (data not shown). This 18 bp nucleotide deletion results in a six-amino-acid deletion in the CD2v of ASF/VN/CanTho-OM/2021 (Figure 2).
Figure 1.
A maximum-likelihood phylogenetic tree based on the complete sequences of the EP402R gene (encoding for serotype-specific proteins CD2v) of ASFV. The Kimura 2-parameter model was used to construct the phylogenetic tree using MEGA 7.0. The numbers along the branches indicate bootstrap values of >70% (1000 replicates). The bars and numbers on the right indicate the ASFV serotypes. Black circles indicate the ASFV detected in this study, ASF/VN/CanTho-OM/2021, which caused an outbreak in the Mekong Delta in 2021.
Figure 2.
Alignment of the partial sequences in the EP402R of ASF/VN/CanTho-OM/2021 and other reference ASFVs showing an 18 bp nucleotide deletion of ASF/VN/CanTho-OM/2021. The top panel shows the chromatogram trace from Sanger sequencing for the partial nucleotide sequence containing the 18 bp nucleotide deletion in the EP402R gene of ASF/VN/CanTho-OM/2021. The black triangle indicates the deletion position.
Similarly, ASF/VN/CanTho-OM/2021 has a repeated insertion of 78-nucleotide sequence in the B475L gene “GAAGAAACTATTCTTGCGATTAAACAGGATATATCTGAAGAAGATAATATTTTTGCGATTGATCAGGATAAACCTGAG” at positions 91,352 to 91,356 corresponding to 26-amino-acid insertion (Figure 3). The repeated insertion was also first observed in GenBank, with no identical sequence previously deposited (data not shown).
Figure 3.
Alignment of the partial nucleotide sequences in the B475L of ASF/VN/CanTho-OM/2021 and other reference ASFVs showing a 78 bp insertion of ASF/VN/CanTho-OM/2021. The underlined and boldfaced characters indicate the original and repeated insertion sequences in the B475L of ASF/VN/CanTho-OM/2021, respectively. The top panel shows the chromatogram trace from Sanger sequencing for the partial nucleotide sequence containing the 78 bp insertion in the B475L gene of ASF/VN/CanTho-OM/2021.
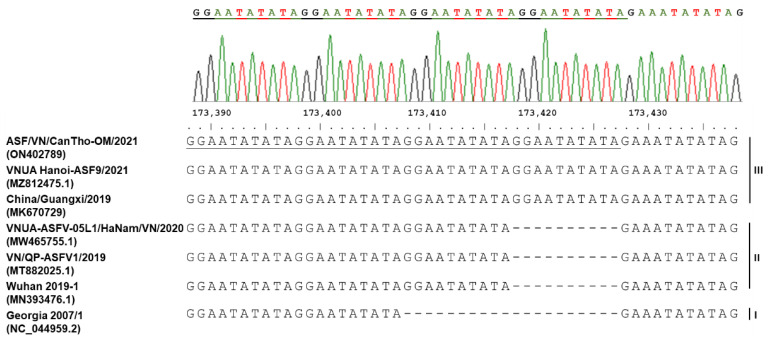
In addition, the nucleotide sequence of the IGR between the I73R and I329L genes of ASF/VN/CanTho-OM/2021 contains four copies of TRS “GGAATATATA”, which are identical to the nucleotide sequence of VNUA/Hanoi-ASF9/2021 (MZ812475.1) and China/Guangxi/2019 (MK670729.1), which all belong to IGR III variant classification (Figure 4). Therefore, ASF/VN/CanTho-OM/2021 can be classified into the IGR III variant of genotype 2.
Figure 4.
Alignment of the partial nucleotide sequences of the IGR between the l73R and I329L genes of ASF/VN/CanTho-OM/2021 and other reference ASFVs indicating four copies of TRS “GGAATATATA” (underlined characters) of ASF/VN/CanTho-OM/2021. The top panel shows the chromatogram trace from Sanger sequencing for the partial nucleotide sequence containing four copies of TRS of ASF/VN/CanTho-OM/2021.
3. Discussion
In Vietnam, the first ASF outbreak was reported in February 2019. The disease quickly spread across all provinces, and ASF is currently endemic all over the country, with serial outbreak reports [10,13]. According to the Vietnam Department of Animal Health, from 1 January 2022 to 29 March 2022, a total of 562 ASF outbreaks were reported from 42 provinces in all northern, central and southern regions (https://www.fao.org/ag/againfo/programmes/en/empres/ASF/situation_update.html, accessed on 1 May 2022).
Owing to the importance and widespread distribution of ASF in Vietnam, many studies have been conducted for the genetic characterization of circulating ASFVs and tracing incursion of viruses based on specific molecular markers [10,11,12]. Most focused on ASFVs detected in the northern region of Vietnam, but limited information on ASFVs circulating in the Mekong Delta, the southernmost region comprising 1 central city and 12 provinces, is available. In this study, a causative ASFV for a fatal outbreak in domestic pigs in the Mekong Delta was subjected to complete-genome analysis. In October 2021, a backyard pig farm suspected with ASF was reported in Can Tho city, which is considered the epicenter of the Mekong Delta. The presence of ASFV was first confirmed using conventional PCR, which targeted the B646L gene. Subsequently, the complete genome of ASFV, ASF/VN/CanTho-OM/2021, was obtained using NGS to better understand the evolution and incursion of ASFV in the region.
The phylogenetic analysis based on B646L (p72) and EP402R (CD2v) sequences indicated that ASF/VN/CanTho-OM/2021 belongs to genotype 2 and serotype 8 (Figure 1). This result was consistent with previous reports showing that all ASFVs causing outbreaks in Vietnam belong to genotype 2 and serotype 8 [10,11,12]. This is the most predominant genotype/serotype of invasive ASFVs currently circulating in several Asian and European countries, all derived from the invasive Georgia/2007/1 isolate [15].
Intriguingly, an 18 bp nucleotide deletion of “CTACTACCCAATATCCCG” was detected in the EP402R gene, which resulted in a six-amino-acid deletion “LLPNIP” in the CD2v of ASF/VN/CanTho-OM/2021 (Figure 2). The short mutation sequence in the EP402R was confirmed as unique because it has no homology to other sequences previously deposited in GenBank (data not shown). Nevertheless, this mutation was not detected in the phylogenetic tree of EP402R because gaps resulting from a deletion in the DNA alignment were treated as missing data; therefore, it was not accounted for in statistical estimation in common phylogenetic analyses. A previous study also argued that phylogenetic analysis of several important genes of ASFV, such as B646L (p72) and EP402R (CD2v), did not accurately define the genotypes and serotypes of ASFVs [16]. Thus, this study remains limited as it did not fully elucidate how the deletion affects the sero-characteristics of the virus.
Based on complete-genome analysis, this study detected another unique mutation in the B475L that had a repeated insertion of the 78-nucleotide sequence in the B475L gene “GAAGAAACTATTCTTGCGATTAAACAGGATATATCTGAAGAAGATAATATTTTTGCGATTGATCAGGATAAACCTGAG” (Figure 3). In fact, the function of B475L remains unknown. Therefore, further studies and information are needed to determine the role of the gene and the causation of the mutation in the B475L to improve the understanding of the pathogenicity and evolution of ASFVs. On the other hand, it is believed that ASF/VN/CanTho-OM/2021 is highly virulent in domestic pigs because infected animals display obvious clinical symptoms of acute ASF infection, such as hyperthermia, respiratory distress, skin hemorrhages, hemorrhagic splenomegaly and lymph node and multiorgan hemorrhages.
The sequence of TRS insertions in IGR between the I73R and I329L genes is widely used as a genome marker to discriminate closely circulating ASFVs worldwide, including in Vietnam [14,17,18]. The comparative DNA alignment based on the IGR between the I73R and I329L genes showed that TRS insertions of ASF/VN/CanTho-OM/2021 are similar to other ASFVs detected in northern Vietnam and China (Figure 4). It is assumed that the widespread distribution of the IGR III variant is a result of the incursion of ASFVs in China to several other Asian countries, including Vietnam [14,17,18]. Currently, at least three different variants of ASFV (IGR I, II and III) circulate in the domestic pig population in Vietnam [13]. Detection of circulation of the IGR III variant might provide important information for the incursion of ASFV in the Mekong Delta, which is more likely from the northern to southern regions in Vietnam [12,13]. It is also noteworthy that no nucleotide mutation was found in the other major functional genes of ASF/VN/CanTho-OM/2021, such as O61R (p12, attachment protein), CP204L (p30, phosphoprotein involved in virus entry) and E183L (p54, binds to the LC8 chain of dynein involved in virus entry; Supplemental Figure S1).
This study is the first to report the complete genome of ASFV in the Mekong Delta and detect a distinct variant of genotype 2. The disease remains highly endemic in the region, but little information is known about circulating viruses. Therefore, this study provides great significance for understanding the epidemiology, evolution and incursion of ASFV in the Mekong Delta and Vietnam.
4. Materials and Methods
4.1. Sample Collection and Sampling Site
This study was conducted following a fatal virus outbreak in domestic pigs on a backyard farm in Can Tho city, a central administrative division of the Mekong Delta with a distance of about 1500 km from Ha Noi capital, in October 2021. All 11 infected pigs displayed obvious clinical symptoms of ASF infection. The samples were collected from organs (spleen and lymph nodes) of 2 dead pigs out of the 11 clinically infected pigs at the farm and used for conventional PCR targeting of the B646L gene to confirm the presence of ASFV, as described previously [5].
4.2. Whole-Genome Sequencing
NGS was used to obtain the WGS of the detected ASFV. Briefly, total viral DNAs were directly extracted from the collected organs using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA). The WGS was performed at the KTest Science Company (Ho Chi Minh City, Vietnam) using the Illumina MiniSeq platform (2 × 150 bp paired ends). Adapters, primers and low-quality sequences (average score < 20 and read length < 35 bp) were removed using fastp version 0.20.1 [19]. The trimmed reads were mapped to the reference genome Sscrofa11 from Ensembl (GCA_000003025.6) using bowtie2 version 2.4.2 [20] with default settings. The unmapped reads were de novo assembled using Unicycler version 0.4.8 [21] with default settings to generate ASFV genomes. Geneious Prime 2021.2.2 was used for scaffolding and annotation of the assembled ASFV genome using Georgia 2007/1 (GenBank accession number NC_044959.2) and Wuhan/2019 (GenBank accession number MN393476) as reference genomes.
4.3. DNA Alignment and Phylogenetic Analysis
The complete genome in a single contig was submitted for downstream analyses such as DNA multiple alignments, trimming and phylogenetic tree construction. The full-length genome sequence of ASFV detected in this study and other ASFV reference strains obtained from GenBank were aligned using MAFFT version 7 with default settings [22]. The quality of the alignment was manually inspected using BioEdit version 7.0.544. A maximum-likelihood tree for each B646L and EP402R sequence was constructed using MEGA 7.0 with a resampling process of 1000 replicates. The manipulation of the DNA sequence was carried out using R version 3.4.4 with the contributed packages ape [23], ips [24] and msa [25].
4.4. PCR Amplification and Sanger Sequencing
Sequences in the genome contig of ASF/VN/CanTho-OM/2021 containing a mutation comparable to the Georgia 2007/1 genome were amplified by conventional PCR and sequenced using the Sanger method for verification. DNA alignments and chromatogram traces from Sanger sequencing were manually inspected using BioEdit version 7.0.5 [26].
Acknowledgments
The authors sincerely thank Takahiro Hiono (Laboratory of Microbiology, Department of Disease Control, Faculty of Veterinary Medicine, Hokkaido University) for his thoughtful reviews and comments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens11070797/s1, Figure S1: Alignments of the full nucleotide sequences of (A) O61R gene (p12, attachment protein); (B) CP204L gene (p30, phosphoprotein involved in virus entry) and (C) E183L gene (p54, binds to the LC8 chain of dynein involved in virus entry) of ASF/VN/CanTho-OM/2021 and other reference ASFVs.
Author Contributions
Conceptualization, N.D.H., L.T.N., N.I. and Y.S.; methodology, N.D.H., L.T.N. and L.T.H.; software, L.T.N., N.N.B. and T.M.Q.; validation, N.D.H., N.I. and Y.S.; formal analysis, L.T.N., L.T.H., N.N.B. and T.M.Q.; investigation, L.T.N., L.T.H., N.N.B. and T.M.Q.; resources, N.D.H., N.I. and Y.S.; writing—original draft preparation, L.T.N., L.T.H. and T.M.Q.; writing—review and editing, N.D.H., N.I. and Y.S.; visualization, L.T.N., N.N.B. and T.M.Q.; supervision, N.D.H., N.I. and Y.S.; project administration, N.D.H. and Y.S.; funding acquisition, N.D.H. and Y.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethical review and approval were waived for this study, because the sampling was a part of the ordinary herd diagnostic and outbreak investigation.
Informed Consent Statement
Not applicable.
Data Availability Statement
The complete-genome sequence generated in this study was submitted to GenBank under accession number: ON402789. Data that support the findings of this study are available upon request from the authors.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The APC was supported in part by the Partnership for Building Resilience against Public Health Emergencies through Advanced Research and Education (PREPARE) project organized by the Japan International Cooperation Agency (JICA).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Costard S., Wieland B., de Glanville W., Jori F., Rowlands R., Vosloo W., Roger F., Pfeiffer D.U., Dixon L.K. African swine fever: How can global spread be prevented? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:2683–2696. doi: 10.1098/rstb.2009.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sánchez-Vizcaíno J.M., Mur L., Martínez-López B. African swine fever: An epidemiological update. Transbound. Emerg. Dis. 2012;59:27–35. doi: 10.1111/j.1865-1682.2011.01293.x. [DOI] [PubMed] [Google Scholar]
- 3.Dixon L.K., Chapman D.A., Netherton C.L., Upton C. African swine fever virus replication and genomics. Virus Res. 2013;173:3–14. doi: 10.1016/j.virusres.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Alejo A., Matamoros T., Guerra M., Andrés G. A proteomic atlas of the African swine fever virus particle. J. Virol. 2018;92:e01293-18. doi: 10.1128/JVI.01293-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastos A.D., Penrith M.L., Crucière C., Edrich J.L., Hutchings G., Roger F., Couacy-Hymann E., Thomson G.R. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Arch. Virol. 2003;148:693–706. doi: 10.1007/s00705-002-0946-8. [DOI] [PubMed] [Google Scholar]
- 6.Montgomery R.E. On a form of swine fever occurring in British East Africa (Kenya Colony) J. Comp. Pathol. Ther. 1921;34:159–191. doi: 10.1016/S0368-1742(21)80031-4. [DOI] [Google Scholar]
- 7.Cwynar P., Stojkov J., Wlazlak K. African swine fever status in Europe. Viruses. 2019;11:310. doi: 10.3390/v11040310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.EFSA (European Food Safety Authority) Baños J.V., Boklund A., Gogin A., Gortázar C., Guberti V., Helyes G., Kantere M., Korytarova D., Linden A. Epidemiological analyses of African swine fever in the European Union: (September 2020 to August 2021) EFSA J. 2022;20:e07290. doi: 10.2903/j.efsa.2022.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mighell E., Ward M.P. African swine fever spread across Asia, 2018–2019. Transbound. Emerg. Dis. 2021;68:2722–2732. doi: 10.1111/tbed.14039. [DOI] [PubMed] [Google Scholar]
- 10.Van Phan L., Dae Gwin J., Sun-Woo Y., Hye-Min K., Thi Bich Ngoc T., Thi Lan N., Thi To Nga B., Jinsik O., Joon Bae K., Kwang Myun C., et al. Outbreak of African swine fever, Vietnam, 2019. Emerg. Infect. Dis. 2019;25:1433–1435. doi: 10.3201/eid2507.190303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mai N.T.A., Vu X.D., Nguyen T.T.H., Nguyen V.T., Trinh T.B.N., Kim Y.J., Kim H.J., Cho K.H., Nguyen T.L., Bui T.T.N., et al. Molecular profile of African swine fever virus (ASFV) circulating in Vietnam during 2019-2020 outbreaks. Arch. Virol. 2021;166:885–890. doi: 10.1007/s00705-020-04936-5. [DOI] [PubMed] [Google Scholar]
- 12.Tran H.T.T., Truong A.D., Dang A.K., Ly D.V., Nguyen C.T., Chu N.T., Nguyen H.T., Dang H.V. Genetic characterization of African swine fever viruses circulating in North Central region of Vietnam. Transbound. Emerg. Dis. 2021;68:1697–1699. doi: 10.1111/tbed.13835. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen V.T., Cho K.-h., Mai N.T.A., Park J.-Y., Trinh T.B.N., Jang M.-K., Nguyen T.T.H., Vu X.D., Nguyen T.L., Nguyen V.D. Multiple variants of African swine fever virus circulating in Vietnam. Arch. Virol. 2022;167:1137–1140. doi: 10.1007/s00705-022-05363-4. [DOI] [PubMed] [Google Scholar]
- 14.Tran H.T.T., Truong A.D., Dang A.K., Ly D.V., Nguyen C.T., Chu N.T., Hoang T.V., Nguyen H.T., Dang H.V. Circulation of two different variants of intergenic region (IGR) located between the I73R and I329L genes of African swine fever virus strains in Vietnam. Transbound. Emerg. Dis. 2021;68:2693–2695. doi: 10.1111/tbed.13996. [DOI] [PubMed] [Google Scholar]
- 15.Chen S.-N., Li C.-L., Lin J.-S., Zhai S.-L., Sun M.-F. Diverse African swine fever viruses in China. New Microbes New Infect. 2022;46:100976. doi: 10.1016/j.nmni.2022.100976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malogolovkin A., Burmakina G., Titov I., Sereda A., Gogin A., Baryshnikova E., Kolbasov D. Comparative analysis of African swine fever virus genotypes and serogroups. Emerg. Infect. Dis. 2015;21:312–315. doi: 10.3201/eid2102.140649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge S., Liu Y., Li L., Wang Q., Li J., Ren W., Liu C., Bao J., Wu X., Wang Z. An extra insertion of tandem repeat sequence in African swine fever virus, China, 2019. Virus Genes. 2019;55:843–847. doi: 10.1007/s11262-019-01704-9. [DOI] [PubMed] [Google Scholar]
- 18.Kim S.H., Lee S.I., Jeong H.G., Yoo J., Jeong H., Choi Y., Son K., Jheong W.H. Rapid emergence of African swine fever virus variants with different numbers of a tandem repeat sequence in South Korea. Transbound. Emerg. Dis. 2021;68:1726–1730. doi: 10.1111/tbed.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S., Zhou Y., Chen Y., Gu J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paradis E., Claude J., Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 24.Heibl C., Cusimano N., Krah F.-S., Heibl M.C. Package ‘ips’, version 0.0.11. 2019. [(accessed on 1 May 2022)]. Available online: http://www.christophheibl.de/Rpackages.html.
- 25.Bodenhofer U., Bonatesta E., Horejš-Kainrath C., Hochreiter S. msa: An R package for multiple sequence alignment. Bioinformatics. 2015;31:3997–3999. doi: 10.1093/bioinformatics/btv494. [DOI] [PubMed] [Google Scholar]
- 26.Hall T. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011;2:60–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete-genome sequence generated in this study was submitted to GenBank under accession number: ON402789. Data that support the findings of this study are available upon request from the authors.