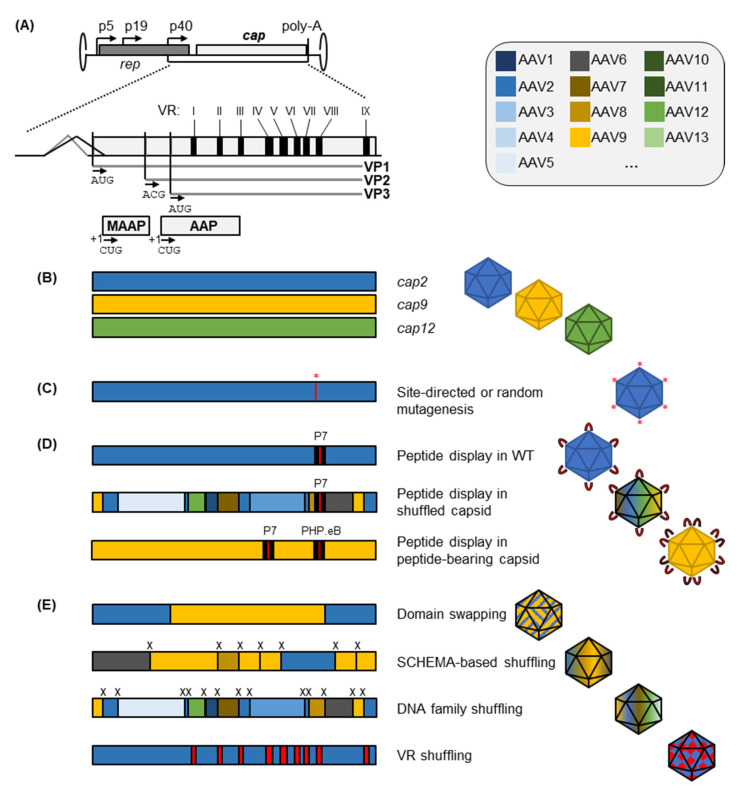
Figure 1.
Structure of the AAV cap gene and technologies for its diversification. (A) Schematic of the AAV cap gene including variable regions (VRs I-IX according to Govindasamy et al. [36]) and transcriptional start sites for VP1, VP2 and VP3, as well as MAAP [37] and AAP [38]. p5, p19 and p40 are the endogenous AAV promoters. poly-A, polyadenylation signal. (B) Tropisms of AAV vectors can be defined by choosing one of 13 primate AAV serotypes (AAV1-13) or a plethora of other naturally occurring isolates from various species. (C) Wild-type tropisms can be modified by mutagenesis of one or several capsid residues (e.g., Kern et al. [39]). (D) Insertion of pre-defined or randomized peptide sequences (e.g., a randomized 7 mer peptide “P7”; red indicates the peptide sequence and black the flanking residues, such as glycine or alanine that can be used as linkers) can be performed within WT cap backbones (e.g., Müller et al. [40]), in synthetic capsids such as shuffled variants (e.g., Tan et al. [41]), or in backbones already carrying an independent peptide insertion in another position (e.g., Goertsen et al. [42]). The colors of the individual capsid fragments denote the serotype origin according to the legends in the upper right corner of this figure. (E) Recombination of larger cap stretches from several parental capsids can be performed via domain swapping (e.g., Shen et al. [43]), SCHEMA-based shuffling through pre-defined optimal crossover points (marked with “x”) (e.g., Ojala et al. [44]), DNA family shuffling based on partial sequence homology (e.g., Grimm et al. [45]), or virtual VR shuffling (e.g., Marsic et al. [46]).