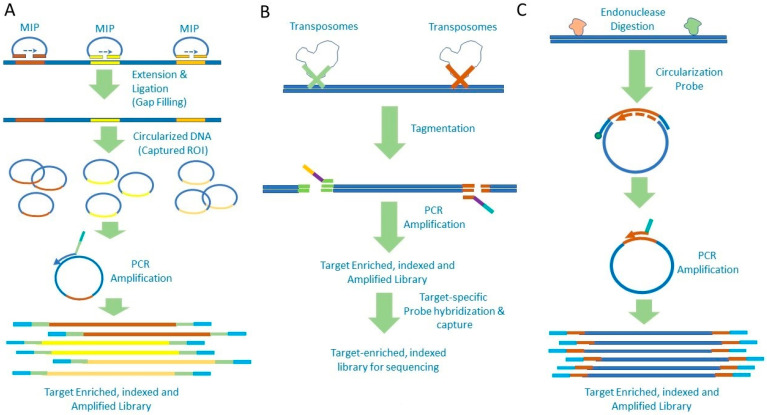
Figure 2.
Variations of the Hybrid Capture Target Enrichment Methods. Some examples of variations of the hybrid capture methods are depicted. (A) Molecular inversion probe-based enrichment, where probes with sequences complementary to the target region of interest are used. Binding of the probes, followed by extension ligation results in circularized DNA with the target region included. Using these as templates, subsequently, a universal primer is used to linearize, amplify, and generate the sequencing library. (B) Tagmentation method, where transposomes are used to fragment double-stranded DNA and tag with sequencing adapters in a single step. This tagged genomic library is amplified using adapter-specific primers followed by hybrid capture with target-specific probes to isolate target-enriched library for sequencing. (C) Haloplex method, where biotin-labeled circularization probes are used to bind to endonuclease-digested DNA, followed by extension ligation, which results in circularized DNA with targeted region of interest, which is amplified using a universal primer to generate enriched library for sequencing.