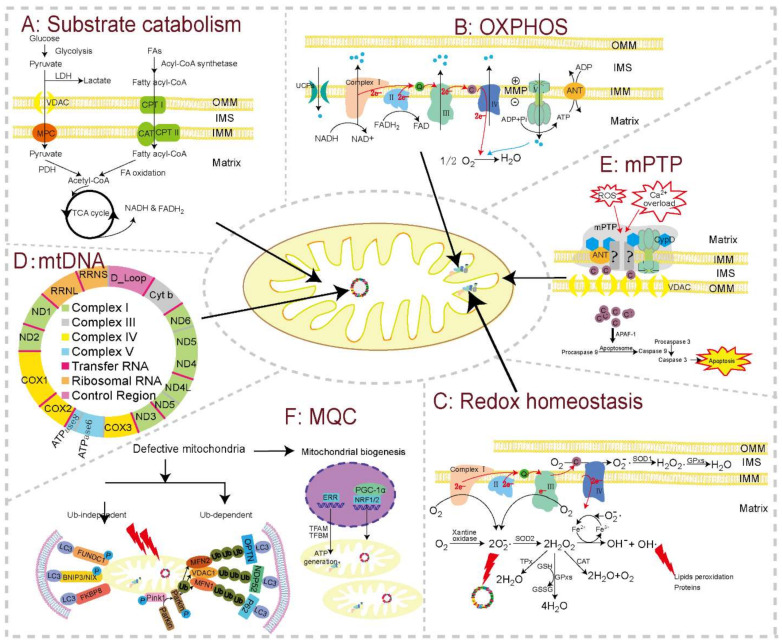
Figure 2.
The basic modules of mitochondrial function and major toxicity targets in cardiomyocytes. Substrate catabolism, oxidative phosphorylation (OXPHOS), redox homeostasis, mitochondrial genome (mtDNA), mitochondrial permeability transition pore (mPTP), and mitochondrial quality control (MQC) constitute the major functional units and toxicity targets in cardiomyocytes. (A) Substrate catabolism. Fatty acids, esterificated by fatty acyl-CoA synthase enzymes, are taken up through CPT I, CPT II, and CAT, and are then oxidized into acetyl-CoA inside the mitochondrion. Pyruvate from glycolysis is also oxidized into acetyl-CoA by PDH in the mitochondrion. Acetyl-CoA then goes through TCA cycle to generate reducing equivalents (NADH and FADH2), which fuel OXPHOS to produce ATP. This bioenergetic process can be disturbed by downregulated expression or decreased activities of carriers and enzymes for the TCA cycle and fatty acids β-oxidation. (B) OXPHOS. Electrons are extracted by complex Ⅰ and II from reducing equivalents, and move through ETC complexes, pumping protons into the IMS to generate MMP. MMP in turn drives proton flow back into the mitochondrial matrix through complex V, releasing this energy to generate ATP. Mitochondrial toxicants can reduce the expression and/or activity of ETC complexes, uncouple ETC from ATP synthesis, and impair MMP. (C) Redox homeostasis. mtROS produced in physiological state can be cleared by series antioxidant enzymes, such as GSH, SOD, and CAT. Drugs with mitochondrial toxicity can overproduce mtROS by inhibiting ETC complexes (especially complex Ⅰ and III) or decreasing the levels or activity of antioxidant enzymes, or there may be a combination of both mechanisms. (D) Map of mtDNA. The mitochondrion possesses its own genome, mtDNA, within the matrix, which can be replicated, transcribed, and translated into some of the MRC complexes. mtDNA, topoisomerase II for mtDNA repair, and DNA polymerase for mtDNA replication are all reported targets for toxicants. (E) mPTP. mPTP is a channel whose components have not been fully elucidated. The normal closed state can be triggered into an open state by a series of stresses, especially Ca2+ overload and oxidative stress. mPTP opening induces cytochrome c releasing into the cytoplasm, resulting in the initiation of apoptosis. (F) MQC. Defective mitochondria can be regulated by MQC, mainly including mitochondrial biogenesis and mitophagy. Damaged mitochondria can be cleared by mitophagy though ubiquitin-dependent or -independent pathways. In cardiomyocytes, ubiquitin-dependent pathway (Pink1-Parkin-mediated mitophagy) is induced by MMP depolarization, while the ubiquitin-independent pathway can be directly induced by LIR containing mitophagy receptors located on OMM in cardiomyocytes. Energy depletion after mitophagy activates genome-encoded transcriptional elements, which directly express mitochondrial proteins or regulate mtDNA to express related proteins for the assembly of new mitochondria. Toxicants may influence mitophagy or biogenesis to disturb MQC. Abbreviations: ANT: adenine nucleotide transporter; APAF: apoptotic peptidase activating factor; BNIP3: BCL2 interacting protein 3; C: cytochrome c; CAT: catalase; CPT: carnitine palmitoyltransferase; Complex I: NADH dehydrogenase; Complex II: succinate dehydrogenase; Complex III: cytochrome c reductase; Complex IV: cytochrome c oxidase; Complex V: ATP synthase; CypD: cyclophilin D; ERR: estrogen-related receptor; ETC: electron transport chain; FAs: fatty acids; FADH2: flavin adenine dinucleotide. FUNDC1: FUN14 domain-containing protein 1; FKBP8: FK506 binding protein 8; GPxs: glutathione peroxidase; GSH: glutathione; GSSG: glutathione disulfide; IBM: inner boundary membrane; IFMs: interfibrillar mitochondria; IMM: the inner mitochondrial membrane; IMS: intermembrane space; LC3: light chain 3; LDH: lactic dehydrogenase; LIR: LC3-interacting region; MFN1/2: mitofusin 1/2; MMP: mitochondrial 6membrane potential; MPC: mitochondrial pyruvate carrier; mPTP: mitochondrial permeability transition pore; NADH: nicotinamide adenine dinucleotide; NRFs: nuclear respiratory factors; OMM: outer mitochondrial membrane; OXPHOS: oxidative phosphorylation; PCMs: primary cardiomyocyte; PDH: pyruvate dehydrogenase; PGC-1α: peroxisome proliferator-activated receptorγ (PPARγ) coactivator 1α; Q: coenzyme Q; SOD: superoxide dismutase; SOD1: Cu/ZnSOD, copper- and zinc-dependent SOD; SOD2: MnSOD, manganese-dependent SOD; SSMs: subsarcolemmal mitochondria; TCA cycle: tricarboxylic acid cycle; TPx: thioredoxin peroxidase; UCP: mitochondrial uncoupling proteins; VDAC: voltage-dependent anion channel.