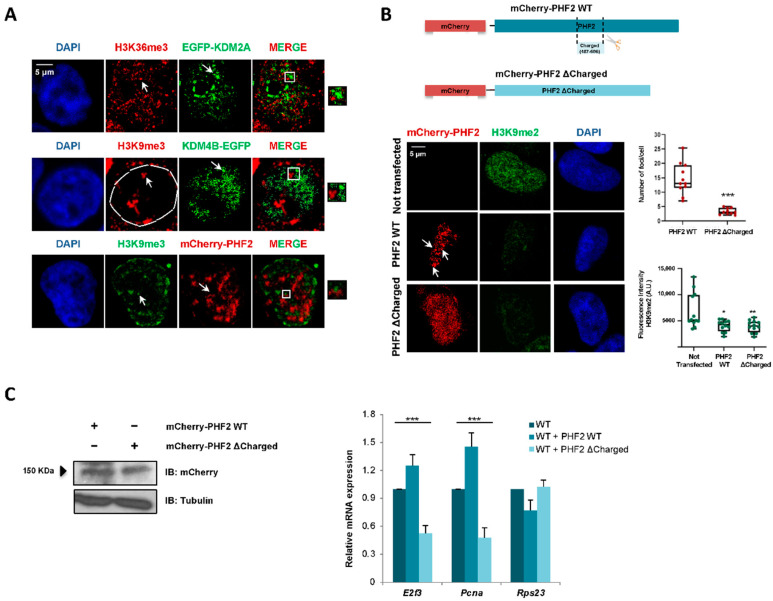
Figure 5.
KDM2A, KDM4B, and PHF2 condensates correlate with transcription. (A) HEK293T cells were transfected with 0.05 μg of EGFP-KDM2A, KDM4B-EGFP, and mCherry-PHF2, and the localization of the active (H3K36me3) or the repressive (H3K9me3) histone marks were analyzed using immunofluorescence staining with anti-H3K36me3 or anti-H3K9me3. The JMJC-KDMs localization was determined following the fluorescence signal. Nuclei were visualized with DAPI (blue). Scale bar, 5 μm. The images are representative of three independent experiments with similar results. (B) mCherry-PHF2 WT and mCherry-PHF2 ΔCharged expression vectors (top panel) were transfected into HEK293T cells. The formation of nuclear puncta was analyzed by following the fluorescence signal. The levels of H3K9me2 were determined by immunofluorescence using anti-H3K9me2 antibody. The boxplots on the right show the number of foci per cell and the H3K9me2 fluorescence intensity. n = 30 transfected cells for condition. Data show the mean ± SEM. For the number of foci per cell, statistic comparisons were made between cells overexpressing PHF2 WT and cells overexpressing PHF2 ΔCharged (*** p < 0.001, Student’s t-test). For fluorescence intensity, comparisons were performed between not transfected cells and cells overexpressing PHF2 WT (* p < 0.05, Student’s t-test) and between not transfected cells and cells overexpressing PHF2 ΔCharged (** p < 0.01, Student’s t-test). Images are representative of 3 biologically independent experiments. Scale bar, 5 μm. (C) PHF2 WT and PHF2 ΔCharged were transfected. 24 h later, total protein extracts were prepared, and the expression levels of PHF2 WT and the mutant were determined by immunoblot using anti-mCherry antibody. The image shown is representative of two independent experiments (left). The expression levels of E2f3 and Pcna genes were quantified by qPCR in NIH3T3 cells (WT, dark blue) expressing PHF2 WT (WT + PHF2 WT, blue) or PHF2 ΔCharged (WT + PHF2 ΔCharged, light blue) (right). As a negative control, the mRNA levels of the housekeeping gene Rps23 were measured. Error bars represent SEM. Statistic comparisons were made between the three groups by means of one-way ANOVA tests (*** p < 0.001). Results are representative of three biologically independent experiments.