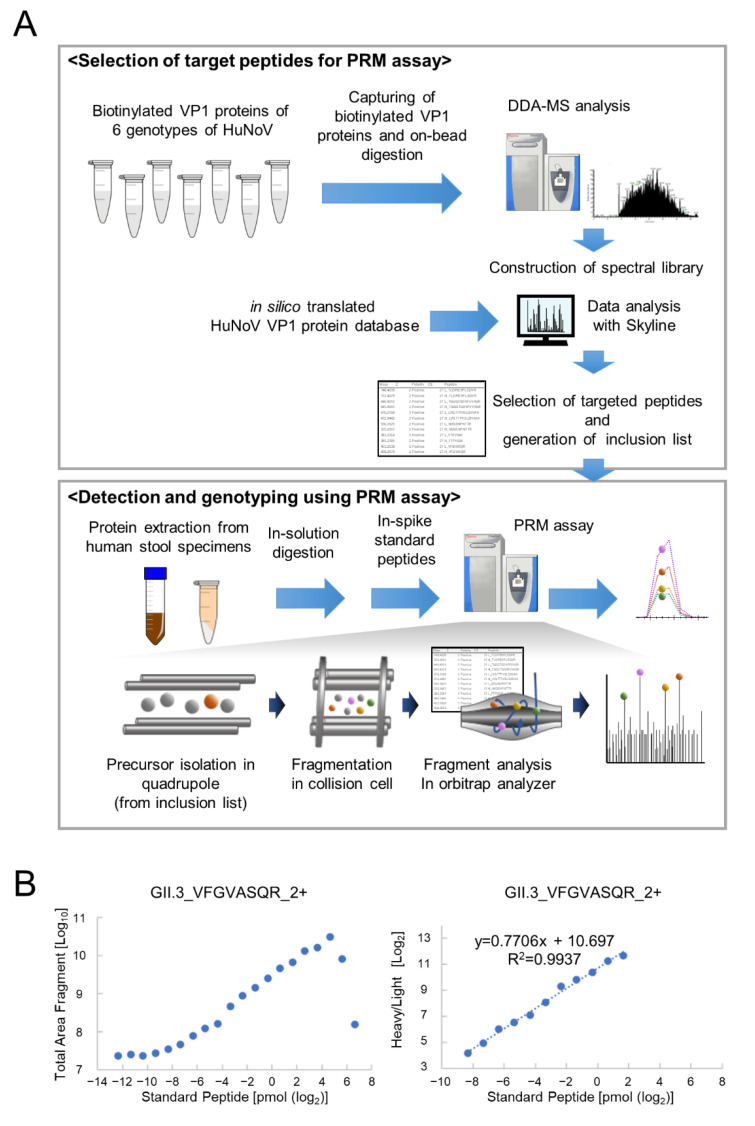
Figure 1.
PRM assay construction for detection and genotyping of the virus. (A) Experimental workflows for PRM assay construction for HuNoV VP1 detection. The peptide information of recombinant HuNoV VP1 proteins was obtained through DDA-MS analysis and in silico translation from the VP1 nucleotide sequence database. The retention time of each heavy isotope-labeled synthetic peptide and the fragment ions’ rank in the PRM assay were analyzed using the Skyline software. Target peptides from the inclusion list were isolated and fragmented from the sample on the basis of the mass-to-charge ratio, and all fragments were analyzed in parallel on a high-resolution mass spectrometer. (B) Example of a calibration curve. The calibration curve was created using the total area fragment of peptide ions. The horizontal axis represents the quantity of standard peptides injected; the vertical axis represents the total area fragment of heavy isotope-labeled peptide ion (left), and the light/heavy ratio (right) obtained using the PRM assay.